作物杂志,2022, 第4期: 37–45 doi: 10.16035/j.issn.1001-7283.2022.04.006
基于转录组数据的生姜ZoWRKY基因家族鉴定及逆境响应分析
姜玉松1,2( ), 李洪雷2, 李哲馨2, 徐晓玉1, 李隆云3, 黄孟军1,2,3(
), 李洪雷2, 李哲馨2, 徐晓玉1, 李隆云3, 黄孟军1,2,3( )
)
- 1西南大学资源环境学院,400715,重庆
2重庆文理学院特色植物研究院,402160,重庆
3重庆市中药研究院中药生药研究所,400065,重庆
Identification and Expression Analysis of the ZoWRKY Family in Stress Responses Based on Transcriptome Data of Ginger (Zingiber officinale Roscoe)
Jiang Yusong1,2( ), Li Honglei2, Li Zhexin2, Xu Xiaoyu1, Li Longyun3, Huang Mengjun1,2,3(
), Li Honglei2, Li Zhexin2, Xu Xiaoyu1, Li Longyun3, Huang Mengjun1,2,3( )
)
- 1College of Resources and Environment, Southwest University, Chongqing 400715, China
2Institute for Special Plants, Chongqing University of Arts and Sciences, Chongqing 402160, China
3Institute of Chinese Materia Medica, Chongqing Academy of Chinese Materia Medica, Chongqing 400065, China
摘要:
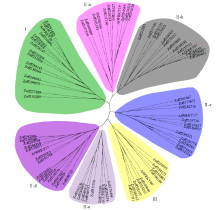
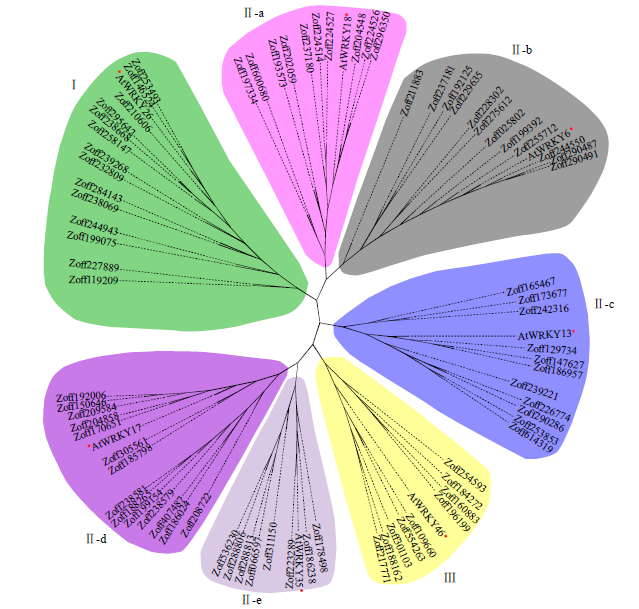
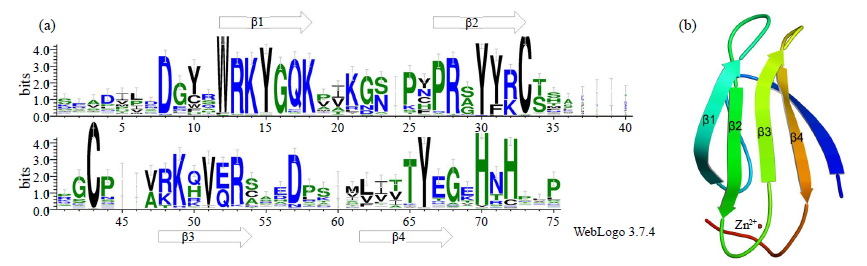
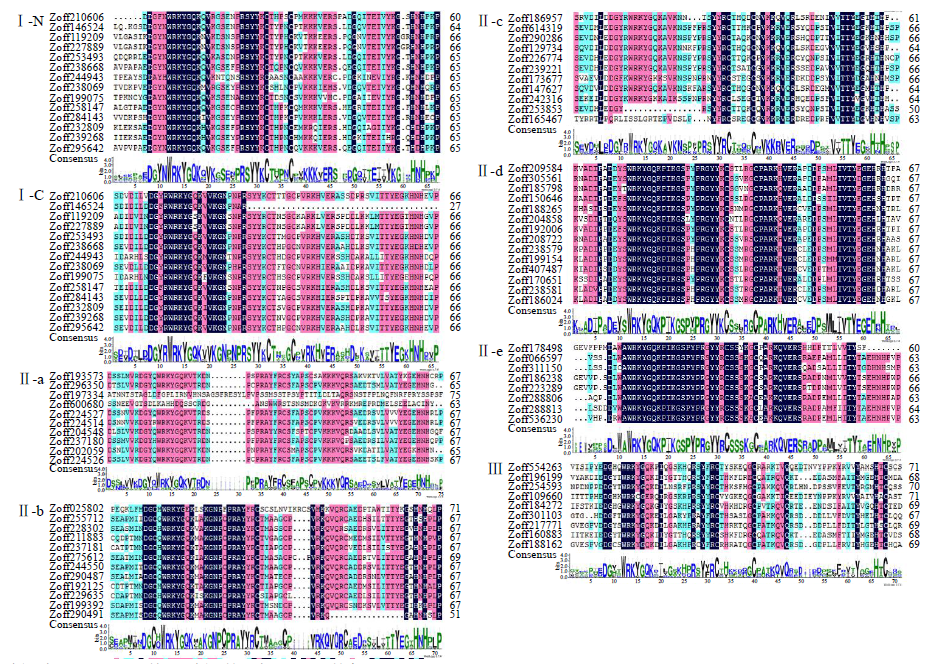
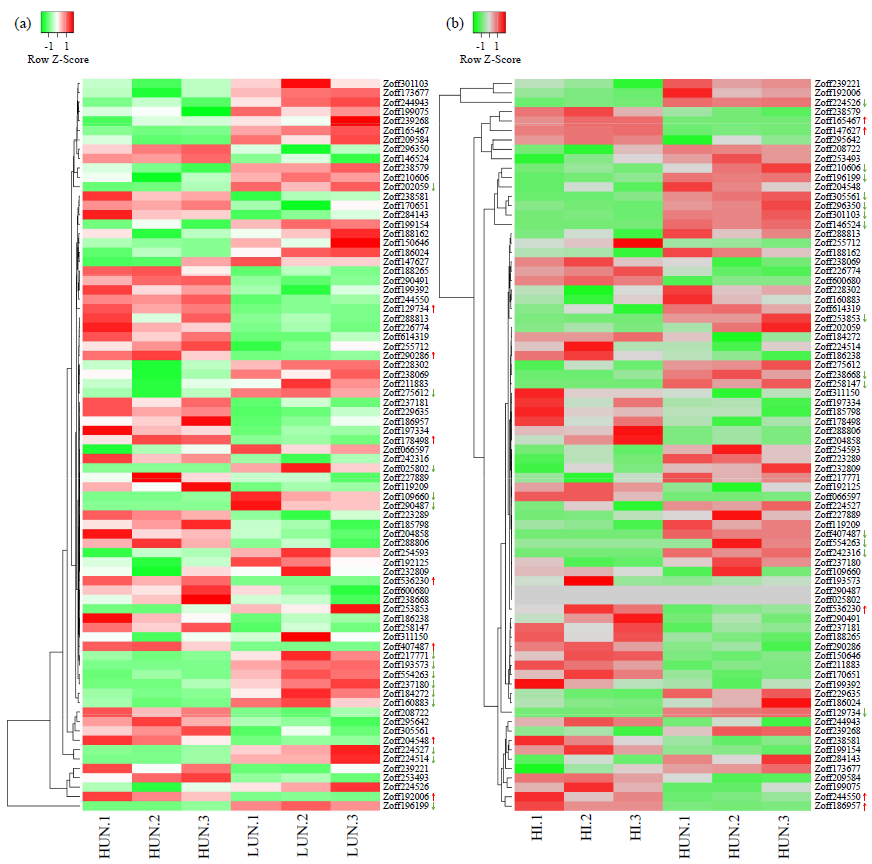
WRKY是一类植物特有的转录因子家族,每个成员都含有高度保守的WRKY结构域,广泛参与植物的生长发育、信号传导和逆境胁迫应答等过程。本研究基于生姜转录组数据鉴定了ZoWRKY家族成员。结果显示,在340.2Mb的生姜转录组数据中,共获得78个具有ORF序列的ZoWRKY家族成员,分为Ⅰ、Ⅱ和Ⅲ 3个亚族,其中亚族Ⅱ又分为Ⅱ-a、Ⅱ-b、Ⅱ-c、Ⅱ-d和Ⅱ-e。逆境响应表达分析显示,分布在Ⅱ-c、Ⅱ-d和Ⅲ等的21个ZoWRKY响应土壤湿度,而分布在Ⅰ、Ⅱ-a和Ⅱ-c等的19个ZoWRKY响应青枯菌侵染,推测ZoWRKY在生姜逆境响应中具有重要功能。对后续开展生姜WRKY的研究、促进抗性育种和提高产量具有重要意义。
| [1] |
Allan A C, Hellens R P, Laing W A. MYB transcription factors that colour our fruit. Trends in Plant Science, 2008, 13(3):99-102.
doi: 10.1016/j.tplants.2007.11.012 |
| [2] |
Liu J, Osbourn A, Ma P. MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Molecular Plant, 2015, 8(5):689-708.
doi: 10.1016/j.molp.2015.03.012 |
| [3] |
Welling A, Palva E T. Involvement of CBF transcription factors in winter hardiness in birch. Plant Physiology, 2008, 147(3):1199-1211.
doi: 10.1104/pp.108.117812 |
| [4] |
Zhao C, Zhang Z, Xie S, et al. Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis. Plant Physiology, 2016, 171(4):2744-2759.
doi: 10.1104/pp.16.00533 |
| [5] |
Olsen A N, Ernst H A, Leggio L L, et al. NAC transcription factors:structurally distinct,functionally diverse. Trends in Plant Science, 2005, 10(2):79-87.
doi: 10.1016/j.tplants.2004.12.010 |
| [6] | Zhang Z, Dong J, Ji C, et al. NAC-type transcription factors regulate accumulation of starch and protein in maize seeds. Proceedings of National Academy of Science of USA, 2019, 116(23):11223-11228. |
| [7] |
黄幸, 丁峰, 彭宏祥, 等. 植物WRKY转录因子家族研究进展. 生物技术通报, 2019, 35(12):129-143.
doi: 10.13560/j.cnki.biotech.bull.1985.2019-0626 |
| [8] |
Wu K L, Guo Z J, Wang H H, et al. The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Research, 2005, 12(1):9-26.
doi: 10.1093/dnares/12.1.9 |
| [9] |
Rushton P J, Somssich I E, Ringler P, et al. WRKY transcription factors. Trends in Plant Science, 2010, 15(5):247-258.
doi: 10.1016/j.tplants.2010.02.006 pmid: 20304701 |
| [10] |
Eulgem T, Rushton P J, Robatzek S, et al. The WRKY superfamily of plant transcription factors. Trends in Plant Science, 2000, 5(5):199-206.
pmid: 10785665 |
| [11] | Chen L, Zhang L, Li D, et al. WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proceedings of National Academy of Science of USA, 2013, 110(21):1963-1971. |
| [12] |
Jiang J, Ma S, Ye N, et al. WRKY transcription factors in plant responses to stresses. Journal of Integrative Plant Biology, 2017, 59(2):86-101.
doi: 10.1111/jipb.12513 |
| [13] |
Dang F F, Wang Y N, Yu L, et al. CaWRKY40,a WRKY protein of pepper,plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant,Cell and Environment, 2013, 36(4):757-774.
doi: 10.1111/pce.12011 |
| [14] |
Merz P R, Moser T, Holl J, et al. The transcription factor VvWRKY 33 is involved in the regulation of grapevine (Vitis vinifera) defense against the oomycete pathogen Plasmopara viticola. Physiologia Plantarum, 2015, 153(3):365-380.
doi: 10.1111/ppl.12251 |
| [15] | Cormack R S, Eulgem T, Rushton P J, et al. Leucine zipper-containing WRKY proteins widen the spectrum of immediate early elicitor-induced WRKY transcription factors in parsley. Biochimica Biophysica Acta, 2002, 1576(1):92-100. |
| [16] |
Bakshi M, Oelmuller R. WRKY transcription factors:Jack of many trades in plants. Plant Signaling and Behavior, 2014, 9(2):e27700.
doi: 10.4161/psb.27700 |
| [17] | 徐惠娟, 郑蕊, 陈任, 等. 枸杞WRKY3基因克隆及组织表达分析. 西北植物学报, 2016, 36(9):1721-1727. |
| [18] |
Cai H, Yang S, Yan Y, et al. CaWRKY6 transcriptionally activates CaWRKY40,regulates Ralstonia solanacearum resistance,and confers high-temperature and high-humidity tolerance in pepper. Journal of Experimental Botany, 2015, 66(11):3163-3174.
doi: 10.1093/jxb/erv125 |
| [19] | 李可, 熊茜, 肖晓蓉, 等. 木薯25个WRKY家族转录因子在生物胁迫下的表达分析. 热带生物学报, 2017, 8(1):14-21. |
| [20] |
Ülker B, Somssich I E. WRKY transcription factors:from DNA binding towards biological function. Current Opinion in Plant Biology, 2004, 7(5):491-498.
doi: 10.1016/j.pbi.2004.07.012 |
| [21] |
Ross C A, Liu Y, Shen Q J. The WRKY gene family in rice (Oryza sativa). Journal of Integrative Plant Biology, 2007, 49(6):827-842.
doi: 10.1111/j.1744-7909.2007.00504.x |
| [22] |
Huang S, Gao Y, Liu J, et al. Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Molecular Genetics and Genomics, 2012, 287(6):495-513.
doi: 10.1007/s00438-012-0696-6 |
| [23] |
Dou L, Zhang X, Pang C, et al. Genome-wide analysis of the WRKY gene family in cotton. Molecular Genetics and Genomics, 2014, 289(6):1103-1121.
doi: 10.1007/s00438-014-0872-y |
| [24] |
Semwal R B, Semwal D K, Combrinck S, et al. Gingerols and shogaols:important nutraceutical principles from ginger. Phytochemistry, 2015, 117:554-568.
doi: 10.1016/j.phytochem.2015.07.012 |
| [25] |
Sun C, Palmqvist S, Olsson H, et al. A novel WRKY transcription factor,SUSIBA2,participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. The Plant Cell, 2003, 15(9):2076-2092.
doi: 10.1105/tpc.014597 |
| [26] | 谷彦冰, 冀志蕊, 迟福梅, 等. 苹果WRKY基因家族生物信息学及表达分析. 中国农业科学, 2015, 48(16):3221-3238. |
| [27] |
Eulgem T, Somssich I E. Networks of WRKY transcription factors in defense signaling. Current Opinion in Plant Biology, 2007, 10(4):366-371.
doi: 10.1016/j.pbi.2007.04.020 |
| [28] | 向小华, 吴新儒, 晁江涛, 等. 普通烟草WRKY基因家族的鉴定及表达分析. 遗传, 2016, 38(9):840-856. |
| [29] |
Ali M A, Azeem F, Nawaz M A, et al. Transcription factors WRKY11 and WRKY17 are involved in abiotic stress responses in Arabidopsis. Journal of Plant Physiology, 2018, 226:12-21.
doi: 10.1016/j.jplph.2018.04.007 |
| [30] |
Li J, Brader G, Kariola T, et al. WRKY70 modulates the selection of signaling pathways in plant defense. The Plant Journal, 2006, 46(3):477-491.
doi: 10.1111/j.1365-313X.2006.02712.x |
| [31] |
Chen X, Liu J, Lin G, et al. Overexpression of AtWRKY28 and AtWRKY 75 in Arabidopsis enhances resistance to oxalic acid and Sclerotinia sclerotiorum. Plant Cell Reports, 2013, 32(10):1589-1599.
doi: 10.1007/s00299-013-1469-3 |
| [32] |
Scarpeci T E, Zanor M I, Mueller-Roeber B, et al. Overexpression of AtWRKY 30 enhances abiotic stress tolerance during early growth stages in Arabidopsis thaliana. Plant Molecular Biology, 2013, 83(3):265-277.
doi: 10.1007/s11103-013-0090-8 pmid: 23794142 |
| [33] |
Xu X, Chen C, Fan B, et al. Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40,and WRKY 60 transcription factors. The Plant Cell, 2006, 18(5):1310-1326.
doi: 10.1105/tpc.105.037523 |
| [1] | 王通, 赵孝东, 甄萍萍, 陈静, 陈明娜, 陈娜, 潘丽娟, 王冕, 许静, 禹山林, 迟晓元, 张建成. 花生TCP转录因子的全基因组鉴定及组织表达特性分析[J]. 作物杂志, 2021, (2): 35–44 |
| [2] | 李国龙, 吴海霞, 孙亚卿. 甜菜BvWRKY23基因的RNAi载体构建[J]. 作物杂志, 2020, (5): 41–47 |
| [3] | 徐园园, 赵鹏, 洪权春, 朱晓琴, 裴冬丽. 小麦转录因子基因TaMYB70的分离和表达分析[J]. 作物杂志, 2020, (4): 84–90 |
| [4] | 段俊枝, 齐学礼, 冯丽丽, 张会芳, 孙岩, 燕照玲, 陈海燕, 齐红志, 樊文杰, 杨翠苹, 刘毓侠, 任银玲, 张甲源, 李莹, 卓文飞. 抗旱基因在小麦抗旱基因工程中的应用进展[J]. 作物杂志, 2020, (3): 7–15 |
| [5] | 杨珺凯,沈阳,才晓溪,邬升杨,李建伟,孙明哲,贾博为,孙晓丽. 大豆PHD家族蛋白的全基因组鉴定及表达特征分析[J]. 作物杂志, 2019, (3): 55–65 |
| [6] | 吴昊,李燕敏,谢传晓. 作物耐热生理基础与基因发掘研究进展[J]. 作物杂志, 2018, (5): 1–9 |
| [7] | 张颖,刘朋宇,白雪,杨阳,李玥莹. 黄瓜CsWRKY23基因的生物信息学及表达分析[J]. 作物杂志, 2017, (5): 38–42 |
| [8] | 段俊枝,李莹,赵明忠,李清州,张莉,魏小春,任银铃. NAC转录因子在植物抗非生物胁迫基因工程中的应用进展[J]. 作物杂志, 2017, (2): 14–22 |
| [9] | 邹勇,黄科,姜玉松,刘奕清. 白姜转录组中的SSR位点信息分析[J]. 作物杂志, 2016, (3): 171–174 |
| [10] | 张雪,尹悦佳,范贝,李慧杰,费小钰,崔喜艳. 植物Dof转录因子的结构特点及功能研究进展[J]. 作物杂志, 2016, (2): 14–20 |
| [11] | 李丹丹, 闫丽, 常健敏, 等. 大豆GmWRI1基因在糖、植物激素及盐胁迫下的表达分析[J]. 作物杂志, 2015, (4): 41–46 |
| [12] | 曹士亮, 李文滨, 王石, 等. 农杆菌介导的BcWRKY2基因原位转化玉米茎尖的初步研究[J]. 作物杂志, 2013, (4): 51–56 |
| [13] | 曹士亮, 王成波, 史桂荣, 等. 利用花粉管通道法将BcWRKY2抗旱基因导入玉米的研究[J]. 作物杂志, 2013, (1): 32–36 |
| [14] | 张欢, 张丽莉, 卢翠华, 王石, 潘映雪, 林忠平. 农杆菌介导WRKY Ⅱ基因转化马铃薯抗旱性研究[J]. 作物杂志, 2012, (4): 30–33 |
| [15] | 顾大路, 王伟中, 王红军, 等. 大棚生姜高产优质栽培技术[J]. 作物杂志, 2007, (2): 73–74 |
|
||