水稻(Oryza sativa L.)是中国主要的粮食作物之一,2023年我国早稻总产量达到2817.4万t,单位面积产量达5925.4 kg/hm2[1]。在影响水稻产量的主要农艺性状中,生育期是最重要的性状之一,决定了水稻能否获得适宜和安全的生殖生长期。水稻抽穗期是群体50%的穗器官伸出1~2 cm剑叶的时间阶段,指从播种到稻穗从剑叶伸出所需天数[2]。抽穗期是从营养生长转向生殖生长的关键指标,直接决定了水稻生育期长短,也决定了品种的区域适应性和收割后作物轮作方式。水稻抽穗受光照、温度、水分等环境因素及成花基因、表观遗传和多种信号转导过程等自身遗传特性的共同调控[3]。抽穗调控途径主要有光周期途径、赤霉素途径、自调节途径和春化作用等,其中,光周期途径是调控水稻抽穗的关键途径[4-5]。
光周期对植物生长阶段起着重要的调控作用,是植物感知季节变化以及调整发育阶段的重要信号。水稻光周期可以调节抽穗时间,在不同种植环境和季节中表现出不同的适应性差异。光周期对水稻抽穗的调控是一个复杂的分子网络,涉及多个基因和环境因素的相互作用[6]。本文通过抽穗遗传基础、光周期调控水稻抽穗的主要途径、表观遗传在光周期调节抽穗中的作用以及影响光周期调控水稻抽穗的其他环境因素等4个方面进行综述,为深入理解水稻抽穗调控机理提供参考,并促进水稻品种的改良和生产管理措施的制定。
1 水稻抽穗的遗传基础
植物的生长发育由自身的遗传基础决定,水稻从营养生长转变到生殖生长,完成开花授精过程,实现物种的繁衍与进化。通过不同生态环境的驯化作用,水稻演变出适应不同生长环境的成花特性,包括成花素基因和抽穗相关基因等。
1.1 成花素基因
成花诱导是植物感受环境因子的诱导,启动成花基因表达,触发植物进入生殖生长阶段。在1865年,Sachs[7]提出成花诱导物质的假说,以解释成花诱导与叶片感受光照的关联。Chailakhyan[8]在对苍耳进行串联嫁接和光照诱导试验中发现了传递成花诱导的物质“成花素”(florigen)。前人[9-10]发现,长日照植物与短日照植物互相嫁接后,成花过程对日照长短的适应性更广。到20世纪80年代“成花素假说”趋于完善,明确了成花基因是高度保守的FT,如水稻的Hd3a、番茄的SFT、玉米的ZCN8等,其编码磷脂酰乙醇胺结合蛋白(phosphatidylethanolamine-binding protein,PEBP)。植物叶片在受到信号诱导后,在伴胞细胞内质网上产生PEBP,通过胞间连丝进入筛管,并移动到远端组织[11]。
植物的PEBP家族有3类,分别为MFT-Like、FT-Like和TFL1-Like,FT-Like促进成花,而TFL1- Like抑制成花。水稻的PEBP家族可聚类成6类,I类由OsFTL14和OsFTL8组成,II类包括OsRCN1~OsRCN4,III类包括OsMFT1和OsMFT2,IV类包括OsFTL9、OsFTL10、OsFTL12和OsFTL13,V类包括OsFTL1/FTL1、OsFTL2/Hd3a和OsFTL3/RFT1,VI类包括OsFTL4~OsFTL7和OsFTL11[12-13]。水稻13个OsFTL基因家族中,OsFTL2为Hd3a,已被广泛研究,OsFTL3为RFT1[14]。RFT1和Hd3a的单突变和双突变,在短日照下rft1正常抽穗,rft1-hd3a无法抽穗,而Hd3a转基因抽穗较野生型缩短,Hd3a是短日照下重要的成花基因[15-16]。OsFT-L4~OsFT-L13基因似乎与成花无关[17]。
在成花素转运过程中,有多项调节因子参与作用。其中14-3-3蛋白能够以多种机制与靶蛋白相互作用,调控各种细胞功能和生理过程。水稻14-3-3蛋白家族成员可通过与蛋白激酶、转录因子及其他信号分子结合,参与体内多种信号转导途径的调节,其中重要的功能是参与成花素激活复合体(FAC)的形成,是成花素受体[10]。14-3-3蛋白家族可增强FD/FT的互作,且14-3-3蛋白GF14c直接与Hd3a发生相互作用,阻碍Hd3a向顶端分生组织运输。14-3-3蛋白GF14b或GF14c可直接与OsFD1、Hd3a相互作用,形成稳定的复合物FT-14-3-3-FD(即FAC)。同时,14-3-3蛋白、OsFD1、OsRFT1也可形成三元复合物,促进长日照下成花[18-19]。
1.2 抽穗QTL定位
水稻抽穗期受数量性状基因控制,已报道[20]的水稻抽穗相关基因和数量性状位点(QTL)多达700个。Yamamoto等[21]利用日本晴×Kasalath F2及回交群体,定位到5个水稻抽穗相关的QTL。Lin等[22]进一步精细定位抽穗基因Hd1、Hd3a、Hd3b和Hd5。Monna等[23]发现14个与水稻抽穗相关的QTL,即Hd1~Hd14。Matsoukas等[24]在第6号染色体定位到水稻抽穗基因Hd17。魏和平等[25]在重组自交系(RILs)群体中共检测到11个QTL,6个候选基因在双亲间的表达量差异显著。赵凌等[26]利用高密度Bin遗传图谱定位方法,共检测到15个抽穗的QTL,其中4个可能是新的候选基因。至今,挖掘及注释水稻抽穗的QTL,仍是水稻分子遗传研究的热点,根据Gramene(
1.3 抽穗相关基因
Hd1是重要的抽穗基因,也是水稻中最早被克隆和功能研究的抽穗基因。Hd1的功能受光周期调控,并具有双功能性,参与光周期和成花转换的调节过程[30-31]。短日照条件下,Hd1蛋白激活成花素基因表达,促进了水稻的抽穗,并具备进化保守性。在长日照条件下,Hd1的功能转变为抑制作用,这一转变过程由生物钟调控的Hd1表达与成花素介导的光信号传导共同诱导。研究[32]发现,Hd1在长日照条件下的抑制作用与转录因子DTH8功能存在密切联系。DTH8功能丧失导致Hd1激活Hd3a转录活性,促进水稻抽穗;Hd1与DTH8相互作用形成DTH8-Hd1复合物,促使Hd1抑制Hd3a转录,暗示了Hd1双重功能的转换由长日照中的DTH8介导。
随着植物分子生物学与分子遗传学研究的突飞猛进,更多的水稻抽穗调控基因陆续被发掘与克隆,调控抽穗表达激素的合成与转运、信号蛋白的传导与介导、启动子与非编码区的调控、转录因子或甲基化修饰等多途径、系统性网络图逐渐完善与丰富,基因之间、途径之间既相互独立又协同互作,共同构成了水稻抽穗分子调控的网络系统。
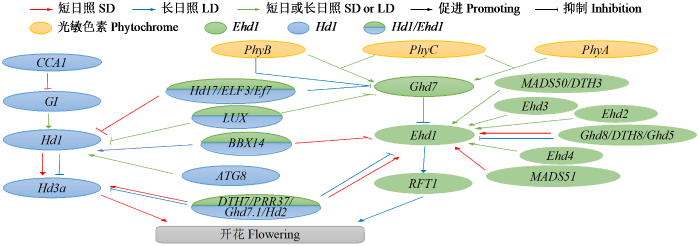
2 光周期调控抽穗的主要途径及参与物质
2.1 光周期调控抽穗的主要途径
图1
图1
光周期调控水稻抽穗的信号网络
Fig.1
The singal pathway regulating heading by photoperiodic sensitive in rice
表1 光周期调控水稻抽穗的主要基因与功能
Table 1
| 基因 Gene | 基因ID Gene ID | 功能 Function | 路径 Pathway | 参考文献 Reference |
|---|---|---|---|---|
| PhyA | Os03g51030 | 光敏色素基因,短日照条件下诱导成花,长日照条件下抑制抽穗 | Ehd1 | [40] |
| PhyB | Os03g19590 | 光敏色素基因,抑制抽穗 | Ehd1 | [41] |
| PhyC | Os03g54084 | 光敏色素基因,抑制抽穗 | Ehd1 | [42] |
| Hd3a | Os06g0157700 | 短日照条件下成花素基因 | Hd1 | [43] |
| RFT1 | Os06g0157500 | 长日照条件下成花素基因 | Ehd1 | [44] |
| GI | Os01g0182600 | 昼夜节律基因,短日照条件下促进抽穗,长日照条件下抑制抽穗 | Hd1 | [45] |
| CCA1/LHY | Os08g0157600 | 昼夜节律基因,具有促进和延迟抽穗的双重功能 | Ehd1/Hd1 | [46] |
| LUX | Os01g74020 | 通过招募OsELF3-1和OsELF4抑制Hd1和Ghd7 | Ehd1/Hd1 | [47] |
| Ehd1 | Os10g0463400 | 长日照条件下的调控抽穗的重要整合因子 | Ehd1 | [33] |
| Ehd2 | Os10g0419200 | 水稻成花的促进因子,调节水稻的成花转变而不影响生长速率 | Ehd1/Hd1 | [48] |
| Ehd3 | Os08g0105000 | 水稻成花的促进因子 | Ehd1 | [49] |
| Ehd4 | Os03g0112700 | 早穗基因,通过Ehd1上调成花素基因的表达从而促进抽穗,但独立于已知的其他Ehd1调控因子 | Ehd1 | [50] |
| Hd1 | Os06g0275000 | 具有双重功能,短日照条件下促进抽穗,长日照条件下抑制抽穗 | Hd1 | [28] |
| Ghd7/Hd4 | Os07g0261200 | 长日照条件下抑制抽穗,Ehd1的主要抑制因子 | Ehd1 | [51] |
| Hd17/ELF3/Ef7 | Os06g0142600 | 长日照条件下,通过抑制Ghd7的表达,上调Ehd1的表达,从而促进抽穗 | Ehd1/Hd1 | [52-53] |
| PRR37/DTH7/ | Os07g0695100 | 长日照条件下,抑制水稻成花素基因并正向调控Ehd1的表达,从而延迟抽穗 | Ehd1/Hd1 | [54] |
| Ghd7.1/Hd2 | ||||
| Ghd8/DTH8/Hd5 | Os08g017450 | 长日照条件下,通过Ehd1介导的途径抑制抽穗 | Ehd1 | [55] |
| MADS51 | Os01g0922800 | 成花促进因子,在短日照下参与OsGI至Ehd1的信号传递 | Ehd1 | [56] |
| MADS50/DTH3 | Os03g0122600 | 长日照条件下促进成花,间接激活Ehd1的表达 | Ehd1 | [57] |
| BBX14 | Os05g0204600 | 成花抑制因子,长日照下促进Hd1表达,作为成花抑制因子发挥作用;短日照下,作为Ehd1的抑制因子发挥作用 | Ehd1/Hd1 | [58] |
| ATG8a-d | Os07g0512200,Os04g0624000,Os11g0100100 | 在短日照和长日照条件下,自噬功能的丧失会导致Hd1的积累并延迟抽穗 | Hd1 | [59] |
2.1.1 OsGI-Hd1-Hd3a途径
2.1.2 Ghd7-Ehd1-Hd3a/RFT1途径
随着抽穗基因的不断挖掘与调控机制深入解析,初步形成光周期调控水稻抽穗的主要通路,参与调控的基因与信号通路,初步勾勒出复杂的信号网络。这2条通路相对独立地发挥作用,但也存在一定联系。如OsGI-Hd1-Hd3a路径的关键基因Hd1能与另一条路径中的Ghd7互作并形成复合体,抑制Ehd1的表达。
2.2 参与水稻抽穗调控的光受体和生物钟基因
已知水稻中有3种光敏色素基因,分别是OsPhyA、OsPhyB和OsPhyC,它们主要感知红光或远红光。OsPhyA和OsPhyB以高度冗余的方式感知持续红光照射,OsPhyC参与感知持续远红光照射[65]。其中,OsPhyA和OsPhyB诱导Ghd7表达,在长日照条件下延迟抽穗,osphyb和osphyc的单突变或双突变都会促进长日照条件下水稻提早抽穗,osphya单突变对水稻抽穗没有影响[42]。Takano等[66]在野生型背景下,组成型表达Ghd7导致延迟抽穗,osphya、osphyb和osphyc的双突变或三突变体,或者光敏色素缺失突变体Se5,长日照诱导不敏感。Zheng等[67]研究表明,光敏色素和OsGI在调节Ghd7蛋白稳定性和抽穗中具有拮抗作用,光敏色素可诱导Ghd7表达以及转录后的调节。光敏色素OsPhyA、OsPhyB和OsGI可以直接与Ghd7相互作用,OsPhyA和OsPhyB可以抑制OsGI和Ghd7之间的拮抗作用。同时,Ghd7-OsGI共表达导致Ghd7蛋白的积累减少,部分缩短野生型的抽穗期,表明光敏色素和OsGI在调节Ghd7蛋白稳定性和开花时间方面发挥着拮抗作用。
生物钟(circadian clock)是生物体进化形成的内源性调控机制,参与调控光周期途径的抽穗基因表达[68]。拟南芥中的CCA1及其同源功能基因LHY与TOC1基因组成植物生物钟中央振荡器的核心循环[38]。与拟南芥中LHY/CCA1同源的水稻生物钟基因OsCCA1/OsLHY,其突变体oscca1/oslhy在长日照下抽穗延迟,而在短日照下抽穗提早,OsLHY的双重功能完全依赖于OsGI-Hd1途径。OsLHY抑制OsGI、Hd1、Ghd7、Hd3a、RFT1和OsELF3的转录,并激活Ehd1的转录[33,46]。水稻OsPRRs家族中的OsPRR73是生物钟中央振荡器基因的上游调控因子,作用下游靶标OsCCA1/OsLHY和Ehd1,OsPRR73与成花基因Ehd1和生物钟基因OsLHY的启动子结合,在白昼时抑制其表达;OsPRR73在osphyb中表达降低,过表达OsPRR73互补osphyb的早抽穗表型,表明OsPhyB是OsPRR73的上游作用因子[69]。水稻有2个ELF3同源基因OsELF3-1和OsELF3-2,OsELF3-1可调控光周期成花基因在白昼下的表达,通过抑制光敏色素信号通路相关基因PhyA、PhyB、PhyC抑制其下游的Ghd7表达[70]。水稻OsELF3-1和OsELF4与生物钟基因OsLUX形成蛋白复合体EC(OsELF4s+OsELF3-1+OsLUX),EC复合体与Hd1和Ghd7启动子中的LBS元件(LUX结合位点)结合,抑制光周期成花基因表达[47]。上述研究揭示了水稻生物钟基因与抽穗基因的信号传导途径的调控和交互作用。
3 光周期调控水稻抽穗基因的表观遗传
表观遗传是指在生物体内非DNA序列变化发生的可遗传变异,包括DNA甲基化(DNA methylation)、组蛋白修饰(histone modification)、染色质重塑(chromatin remodeling)、非编码RNA(non-coding RNA)及小RNA(microRNAs)等类型[59]。表观遗传修饰通过广泛参与光周期调控水稻抽穗。
3.1 DNA甲基化调控抽穗的表观遗传
DNA甲基化是表观遗传的重要修饰形式之一,在植物中可发生在CG、CHG和CHH等位点,受甲基化转移酶和去甲基化酶调控[71]。Kondo等[72]报道了引起DNA去甲基化的胞苷类似物5- Azacytidine(azaC)在非诱导光周期条件下能诱导长日照植物高雪轮(Silene armeria L.)和短日照植物紫苏(Perilla frutescens var. crispa)开花,说明光周期成花相关基因的表达受DNA甲基化调控。Li等[73]报道,OsSAMS1、2和3基因是组蛋白H3K4me3和DNA甲基化调控水稻抽穗相关基因表达所必需的,S-腺苷甲硫氨酸合成酶(S- adenosylmethionine synthase,SAMS)是生物体内重要的甲基供体SAM生物合成途径的一个关键酶。DNA甲基化对水稻抽穗的调控具有一定的影响。RNA干扰下调OsSAMS1、2和3,植株出现矮化、育性降低、发芽和抽穗延迟等表型,外源施用SAM可明显恢复发芽延迟。敲低OsSAMS1、2和3基因抽穗明显推迟,且Ehd1、Hd3a和RFT1的表达下调。
3.2 组蛋白甲基化修饰调控抽穗的表观遗传
Fujino[32]表明,水稻中的DTH8通过DTH8- Hd1模块介导在Hd3a的转录调控中发挥作用,DTH8与Hd3a启动子结合,调节Hd3a基因启动子区的H3K27三甲基化(H3K27me3)水平。DTH8的缺失导致Hd3a启动子区的H3K27me3水平降低,从而实现对光周期成花的表观遗传修饰。Liu等[75]报道了在长日或短日照条件下,突变组蛋白H3K36甲基转移酶SDG708(Set domain group 708)显著降低体内H3K36me1、H3K36me2和H3K36me3的甲基化水平,并延迟水稻抽穗。H3K4特异的甲基转移酶SDG701通过与染色质结合,促进H3K4me3,增强水稻Hd3a和RFT1的表达。Liu等[76]报道,水稻Zeste基因增强组蛋白H3K27甲基转移酶基因SDG711和SDG718,编码PRC2的关键亚基,是组蛋白H3K27me3的必需因子,参与长日和短日照条件下水稻抽穗调控。SDG718下调在长日照下无作用,短日照下延迟抽穗。在短日照条件下SDG718抑制OsLF(Leafy),导致Hd1高表达,从而促进成花转变。SDG711的过量表达在长日照下抑制成花,但在短日照下没有作用。SDG711在长日照下抑制OsLF(Hd1抑制因子),导致Hd1高表达,致使长日照下抑制成花;同时发现SDG711也参与抑制长日照下的Ehd1。
3.3 泛素化修饰调控抽穗的表观遗传
蛋白翻译后的修饰决定了蛋白稳定性和功能活性,研究[81]表明翻译后修饰调节了水稻光周期和抽穗。E3泛素连接酶(heading date associated factor 1,HAF1)通过与Hd1互作并促使其降解影响抽穗,其功能缺失导致抽穗延迟。在长日照条件下,HAF1可能通过对其他成花调节因子的泛素化,调控水稻抽穗。OsELF3是HAF1的直接底物,参与昼夜节律调控,是一个受光周期诱导的抽穗调节因子[81]。DTH2编码促进水稻抽穗的Constans-like蛋白,体外泛素化试验表明HAF1介导DTH2的泛素化,DTH2是HAF1泛素化的直接底物,HAF1通过26S蛋白酶体途径介导DTH2降解[82]。
E3泛素化连接酶(squamosa promoter binding protein-like 11,SPL11)与SPIN1(Spindlin 1)互作,在短日照条件下,介导Hd1抑制成花素基因的转录活性推迟抽穗,而在长日照条件下不影响Hd1表达。SPIN1可与RBS1(RNA-binding and SPIN1- interacting 1)互作,在长日照和短日照下均抑制水稻抽穗。并且,RBS1还可激活SPIN1转录以及抑制SPL11的表达[83]。水稻组蛋白单泛素化基因OsHUB1和2通过Hd1和Ehd1途径参与抽穗调节,在长日照条件下诱导Hd3a、RFT1和Ehd1的表达,且Hd1、Ghd7、OsCCA1、OsGI、OsFKF1和OsTOC1的转录水平降低,而在短日条件下诱导RFT1和Ehd1表达[84]。
4 其他信号调控水稻抽穗
水稻光周期还受其他外源信号的调控,如温度、干旱等。温度感知信号与光周期密切相关,高温可诱导水稻提前抽穗,低温导致水稻抽穗延迟[3]。低温条件下,Ehd1、Hd3a和RFT1表达受到抑制,Ghd7的表达被诱导,导致水稻抽穗延迟[85]。光周期足够时,PhyB作为温度感受器,调控Ghd7抑制蛋白的活性,在低温下抑制Ehd1、Hd3a和RFT1表达;在高温下Ghd7 mRNA减少,导致水稻抽穗基因mRNA的增加。可见,温度响应与光周期响应既独立又交叉互作,协同调控水稻抽穗基因的表达[86]。Hd1在长日照下阻遏蛋白的作用在较低的环境温度下增强,并且hd1突变体中开花对温度变化的敏感性大大降低。来自Koshihikari的OsPRR37等位基因在温度范围内具有相反的作用,当平均环境温度低于临界阈值时,OsPRR37抑制抽穗,但在较高温度下恢复为启动子,促进成花[87]。
植物激素是一种广泛参与植物生长发育的调节物质,其中主要调控因素为赤霉素(GA)和脱落酸(ABA)。GA可与光周期信号协同调控植物的开花,通过光周期中的关键转录因子CO蛋白,GA能够引发成花素基因FT的表达[88]。GA途径的抑制因子DELLA蛋白与CO蛋白相互作用并形成蛋白复合物,从而抑制其转录功能。在开花的诱导过程中,DELLA蛋白仅部分依赖于CO和FT介导的光周期信号通路[89]。Dai等[90]发现EL1是第1个被证实能通过GA诱导途径参与控制水稻抽穗的调控基因。外源的GA3能抑制EL1的表达水平,而EL1能特异磷酸化水稻中的DELLA蛋白SLR1,从而调控GA信号反应和植物的发育,影响水稻抽穗。SLR1是响应水稻GA信号的重要调控蛋白,主要通过水稻中的Ehd1-Hd3a/RFT1路径来负调控抽穗。RID1是水稻成花转换的分子开关,并且编码一种C2H2类锌指转录因子。研究[91]表明,SLR1能够与RID1互作,进而将GA信号途径整合到水稻抽穗调控中。ABA作为植物中重要的激素,参与了调控植物成花。水稻中受ABA诱导表达的OsABF1基因编码bZIP转录因子。研究[92]表明,OsABF1可响应外源ABA诱导,并鉴定“SAPK8-OsABF1-Ehd1/Ehd2”为抑制水稻抽穗的信号通路。
干旱影响植株的生长发育,促使植株由营养生长转向生殖生长,这种生理反应也可称为干旱逃逸(DE)[93]。研究[94]表明,Ehd1、Hd3a和RFT1通过整合光周期和干旱胁迫信号来延迟水稻的成花转变,当遭受干旱胁迫时,会使基因的表达下调,从而推迟水稻抽穗。OsbZIP23受干旱诱导并且正调DE,在干旱处理时,osbzip23突变体呈现延迟抽穗,相反OsbZIP23过表达植株则呈现明显提前抽穗。在干旱条件下,bZIP23反馈抽穗基因的调节,OsTOC1、Ehd1、RID1、Hd3a以及RFT1表达均显著下调,从而推迟水稻抽穗[95]。在干旱条件下,OsABF1基因的表达增强,间接抑制了Ehd1的表达。此外,研究[96]还发现了一种干旱诱导基因OsWRKY104,该基因受OsABF1的直接调控,过表达OsWRKY104可抑制Ehd1表达,并导致水稻开花时间推迟。
5 抽穗基因在水稻改良育种中的应用和展望
中国水稻栽培历史悠久,种植区域广泛,水稻起源于中国南方,而经过人类的长时间驯化栽培,在中国已发展成从南向北多个栽培稻区(18.00~ 53.00°N)。中国水稻研究所根据国内生态环境、社会经济条件和水稻种植方式,将全国划分为六大稻作区:华南双季稻稻作区(18.00~28.00° N),该区位于我国最南部,包括福建、广东、广西等省区,水稻面积占全国的17.7%;华中双季稻稻作区(25.00~35.00° N),含江苏、上海、浙江等省市,占全国水稻面积的68.1%;西南高原单双季稻稻作区(25.00~40.00° N),位于云贵高原和青藏高原,水稻面积占全国7.8%;华北单季稻稻作区(31.00~ 40.00° N),含北京、天津、山东等省市,水稻种植面积占全国的3.3%;东北早熟单季稻稻作区(38.00~53.00° N),包括黑龙江、吉林、辽宁等省,水稻面积占全国2.6%;西北干燥区单季稻稻作区(35.00~49.00° N),包括新疆和宁夏等省区,水稻种植面积仅占全国0.5%[97]。这6个栽培稻区由于纬度和海拔跨度比较大,因此演化生成了不同的水稻感温感光生态型,这也造成水稻跨纬度引种的困难。中国6个水稻生态区抽穗基因上存在着丰富的等位基因变异(表2)。
表2 抽穗基因等位基因型的稻区适应范围
Table 2
| 序号 Number | 稻作区 Rice region | 抽穗基因Heading gene | ||||||
|---|---|---|---|---|---|---|---|---|
| DTH2 | Ehd4 | Hd1 | OsPRR37 | Ghd8 | Ghd7 | |||
| 1 | 华南双季稻稻作区(18~28° N) | 弱等位型 | 弱等位型 | 强等位型或无功能等位型 | 无功能等位型 | 强等位型或无功能等位型 | 强等位型或无功能等位型 | |
| 2 | 华中双季稻稻作区(25~35° N) | 弱等位型或强等位型 | 弱等位型或强等位型 | 强等位型或无功能等位型 | 强等位型或无功能等位型 | 强等位型或无功能等位型 | 强等位型或无功能等位型 | |
| 3 | 西南高原单双季稻稻作区(25~40° N) | 强等位型 | 弱等位型或强等位型 | 强等位型或无功能等位型 | 强等位型或无功能等位型 | 强等位型或无功能等位型 | 强等位型或弱等位型或无功能等位型 | |
| 4 | 华北单季稻稻作区(31~40° N) | 强等位型 | 强等位型 | 强等位型或无功能等位型 | 强等位型无功能等位型 | 无功能等位型 | 弱等位型 | |
| 5 | 东北早熟单季稻稻作区(38~53° N) | 强等位型 | 强等位型 | 无功能等位型 | 无功能等位型 | 无功能等位型 | 弱等位型或无功能等位型 | |
| 6 | 西北干燥区单季稻稻作区(35~49° N) | 强等位型 | 强等位型 | 强等位型或无功能等位型 | 强等位型或无功能等位型 | 无功能等位型 | 弱等位型或无功能等位型 | |
DTH2为效应较小的水稻抽穗数量性状位点,引起抽穗的变化较小,仅在1周内,但对水稻产量和生殖适应具有较大差异;Ehd4为东北早熟单季稻稻作区粳稻地方品种中广泛存在的强功能型等位基因[98]。Ehd4有2个主要单体型,籼稻品系的主要单体型为强功能型Hap2,主要分布在华南双季稻稻作区和华中双季稻稻作区等低纬度和低海拔地区;粳稻品系主要单体型为弱功能型Hap3,主要分布在华中双季稻稻作区、华北单季稻稻作区、东北早熟单季稻稻作区等高纬度和高海拔地区[99]。Hd1基因具有自发的高度多态性,在中国6个主要稻区不同纬度不同海拔地区均发现携带hd1不同等位型的水稻品种,证实Hd1高度多态性[100]。长日照条件下,OsPRR37抑制Hd3a的表达,中国6个主要稻区均存在无功能的PRR37等位基因品种,而功能型等位基因主要存在于华南双季稻稻作区、华中双季稻稻作区、华北单季稻稻作区纬度20~40° N种植区域内[101]。Ghd8在中国存在广泛的变异,携带Ghd8无功能等位基因表现为早抽穗,无功能基因主要分布于高纬度区域,而处于低纬度的品种大部分含有功能型等位基因[42]。强等位型Ghd7效应较大,长日照条件下延迟开花,主要分布在华南双季稻稻作区、华中双季稻稻作区;而无功能型Ghd7分布在华中双季稻稻作区和华南双季稻稻作区以及东北早熟单季稻稻作区中[62]。Ghd8、Ghd7、Hd1与OsPRR37基因大部分变异位点间存在一定的连锁关系,通过人工突变不同组合Hd1、Ghd7、DTH8可以创造出一系列不同抽穗和农艺性状大幅度改变的株系[102]。
随着分子生物学的发展,人们对不同栽培稻区主要控制抽穗的关键基因的单倍型逐渐有了清晰的认识,同时伴随CRISPR-Cas9基因编辑技术的应用发展,在未来生物育种通道上,人们可以运用手术刀将特定的抽穗基因稍加修改,从而加快引种,例如,影响开花时间、株高和每圆锥花序小穗数的多效基因Ghd7,通过敲除可以将强功能型等位基因变弱,从而实现不同纬度引种。Hd1在中国的品种中存在许多等位型,其中粳稻中的等位型变异度没籼稻中变异度高。在长江中下游华中双季稻稻作区的粳稻栽培品种中引入籼稻单倍型H16等位基因,可以延长粳稻生育期,从而实现中粳向晚粳的转变,显著提高粳稻单株产量,并且不会改变与品质相关的性状。编辑DTH8、Ghd7和Hd1启动子中的顺式元件,不同程度地降低了这些基因的表达,削弱了它们对抽穗的抑制作用。在75~107 d的抽穗期获得了突变体材料,从中成功地选择了高产、早抽穗和适应高纬度地区的新种质,以达到理想产量,同时避免霜冻危害[61]。由于抽穗基因强烈影响农艺性状,很多抽穗基因(DTH8、Ghd7、Hd1、Ehd1)已被证明分别对水稻的适应性做出了贡献[103]。然而,单独通过单个基因或多个基因的敲除进行引种带来的正效应仍不明确,许多基因编辑敲除带来的负效应居多,因此在生产上跨生态型引种的应用不多,此外,能使水稻产量最大化的基因组合可能并不存在于自然种群中。因此,进一步开展抽穗多基因的编辑,并在多纬度多海拔试种选育研究将会为以后水稻生物育种提供依据。
参考文献
Environmental control of rice flowering time
Fine-tuning of the setting of critical day length by two casein kinases in rice photoperiodic flowering
DOI:10.1093/jxb/erx412
PMID:29237079
[本文引用: 1]

Many short-day plants have a critical day length that fixes the schedule for flowering time, limiting the range of natural growth habitats (or growth and cultivation areas). Thus, fine-tuning of the critical day-length setting in photoperiodic flowering determines ecological niches within latitudinal clines; however, little is known about the molecular mechanisms controlling the fine-tuning of the critical day-length setting in plants. Previously, we determined that florigen genes are regulated by day length, and identified several key genes involved in setting the critical day length in rice. Using a set of chromosomal segment substitution lines with the genetic background of an elite temperate japonica cultivar, we performed a series of expression analyses of flowering-time genes to identify those responsible for setting the critical day-length in rice. Here, we identified two casein kinase genes, Hd16 and Hd6, which modulate the expression of florigen genes within certain restricted ranges of photoperiod, thereby fine-tuning the critical day length. In addition, we determined that Hd16 functions as an enhancer of the bifunctional action of Hd1 (the Arabidopsis CONSTANS ortholog) in rice. Utilization of the natural variation in Hd16 and Hd6 was only found among temperate japonica cultivars adapted to northern areas. Therefore, this fine-tuning of the setting of the critical day length may contribute to the potential northward expansion of rice cultivation areas.© The Author(s) 2017. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Wirkung des lichtes auf die blütenbilding unter bermittlung der laubblätter
Concerning the hormonal nature of plant development processes
Transmission of the floral stimulus from a long-short-day plant, Bryophyllum daigremontianum, to the short- long-day plant Echeveria harmsii
The physiology of flower initiation in Pisum sativum
Genomic survey of PEBP gene family in rice:Identification, phylogenetic analysis, and expression profiles in organs and under abiotic stresses
The FLOWERING LOCUS T/TERMINAL FLOWER 1 gene family: Functional evolution and molecular mechanisms
DOI:10.1016/j.molp.2015.01.007
PMID:25598141
[本文引用: 1]

In plant development, the flowering transition and inflorescence architecture are modulated by two homologous proteins, FLOWERING LOCUS T (FT) and TERMINAL FLOWER 1 (TFL1). The florigen FT promotes the transition to reproductive development and flowering, while TFL1 represses this transition. Despite their importance to plant adaptation and crop improvement and their extensive study by the plant community, the molecular mechanisms controlling the opposing actions of FT and TFL1 have remained mysterious. Recent studies in multiple species have unveiled diverse roles of the FT/TFL1 gene family in developmental processes other than flowering regulation. In addition, the striking evolution of FT homologs into flowering repressors has occurred independently in several species during the evolution of flowering plants. These reports indicate that the FT/TFL1 gene family is a major target of evolution in nature. Here, we comprehensively survey the conserved and diverse functions of the FT/TFL1 gene family throughout the plant kingdom, summarize new findings regarding the unique evolution of FT in multiple species, and highlight recent work elucidating the molecular mechanisms of these proteins. Copyright © 2015 The Author. Published by Elsevier Inc. All rights reserved.
FT-like proteins induce transposon silencing in the shoot apex during floral induction in rice
Proceedings of the National Academy of Sciences of the United States of America,
Hd3a and RFT1 are essential for flowering in rice
Hd3a protein is a mobile flowering signal in rice
DOI:10.1126/science.1141753
PMID:17446351
[本文引用: 1]

Florigen, the mobile signal that moves from an induced leaf to the shoot apex and causes flowering, has eluded identification since it was first proposed 70 years ago. Understanding the nature of the mobile flowering signal would provide a key insight into the molecular mechanism of floral induction. Recent studies suggest that the Arabidopsis FLOWERING LOCUS T (FT) gene is a candidate for encoding florigen. We show that the protein encoded by Hd3a, a rice ortholog of FT, moves from the leaf to the shoot apical meristem and induces flowering in rice. These results suggest that the Hd3a protein may be the rice florigen.
The significance of florigen activation complex in controlling flowering in rice
Map-based cloning and functional analysis of YE1 in rice, Which is involved in light-dependent chlorophyll biogenesis and photoperiodic flowering pathway
Identification of heading date quantitative trait locus Hd6 and characterization of its epistatic interactions with Hd2 in rice using advanced backcross progeny
DOI:10.1093/genetics/154.2.885
PMID:10655238
[本文引用: 1]

A backcrossed population (BC(4)F(2)) derived from a cross between a japonica rice variety, Nipponbare, as the recurrent parent and an indica rice variety, Kasalath, as the donor parent showed a long-range variation in days to heading. Quantitative trait loci (QTL) analysis revealed that two QTL, one on chromosome 3, designated Hd6, and another on chromosome 2, designated Hd7, were involved in this variation; and Hd6 was precisely mapped as a single Mendelian factor by using progeny testing (BC(4)F(3)). The nearly isogenic line with QTL (QTL-NIL) that carries the chromosomal segment from Kasalath for the Hd6 region in Nipponbare's genetic background was developed by marker-assisted selection. In a day-length treatment test, the QTL-NIL for Hd6 prominently increased days to heading under a 13.5-hr day length compared with the recurrent parent, Nipponbare, suggesting that Hd6 controls photoperiod sensitivity. QTL analysis of the F(2) population derived from a cross between the QTL-NILs revealed existence of an epistatic interaction between Hd2, which is one of the photoperiod sensitivity genes detected in a previous analysis, and Hd6. The day-length treatment tests of these QTL-NILs, including the line introgressing both Hd2 and Hd6, also indicated an epistatic interaction for photoperiod sensitivity between them.
Characterization and detection of epistatic interactions of 3 QTLs, Hd1, Hd2, and Hd3, controlling heading date in rice using nearly isogenic lines
Genetic dissection of a genomic region for a quantitative trait locus, Hd3, into two loci, Hd3a and Hd3b, controlling heading date in rice
DOI:10.1007/s00122-001-0813-0
PMID:12582636
[本文引用: 1]

The rice photoperiod sensitivity gene Hd3 was originally detected as a heading date-related quantitative trait locus localized on chromosome 6 of rice. High-resolution linkage mapping of Hd3 was performed using a large segregating population derived from advanced backcross progeny between a japonica variety, Nipponbare, and an indica variety, Kasalath. To determine the genotype of Hd3, we employed progeny testing under natural field and short-day conditions. As a result, two tightly linked loci, Hd3a and Hd3b, were identified in the Hd3 region. Nearly-isogenic lines for Hd3a and Hd3b were selected from progeny using marker-assisted selection. The inheritance mode of both Hd3a and Hd3b was found to be additive. Analysis of daylength response in nearly-isogenic lines of Hd3a and Hd3b showed that the Kasalath allele at Hd3a promotes heading under short-day conditions while that at Hd3b causes late heading under long-day and natural field conditions.
Florigenic and antiflorigenic signaling in plants
DOI:10.1093/pcp/pcs130
PMID:23008422
[本文引用: 1]

The evidence that FLOWERING LOCUS T (FT) protein, and its paralog TWIN SISTER OF FT, act as the long-distance floral stimulus, or at least that they are part of it in diverse plant species, has attracted much attention in recent years. Studies to understand the physiological and molecular apparatuses that integrate spatial and temporal signals to regulate developmental transitions in plants have occupied countless scientists and have resulted in an unmanageably large amount of research data. Analysis of these data has helped to identify multiple systemic florigenic and antiflorigenic regulators. This study gives an overview of the recent research on gene products, phytohormones and other metabolites that have been demonstrated to have florigenic or antiflorigenic functions in plants.
水稻抽穗期QTL定位及候选基因分析
DOI:10.11983/CBB22114
[本文引用: 1]

水稻(Oryza sativa)抽穗期是决定产量和品质的重要性状, 在育种、制种及引种驯化过程中发挥重要作用。将热研2号(O. sativa subsp. japonica cv. ‘Nekken2’)和华占(O. sativa subsp. indica cv. ‘HZ’)杂交获得F<sub>1</sub>代, 经连续多代自交得到120个重组自交系(RILs)群体。在常规水肥管理条件下, 对120个RILs株系的抽穗时间进行统计分析。利用已构建好的高密度遗传图谱, 对水稻抽穗期相关性状进行QTL定位分析, 结果共检测到11个QTLs, 分别位于第1、3、4、5、6、8和12号染色体上, 其中1个LOD值高达5.75。通过分析QTLs区间内的候选基因, 筛选出可能影响两亲本抽穗期的相关基因, 并利用实时定量PCR进行基因表达量分析, 发现LOC_Os03g03070、LOC_Os03g50310、LOC_Os03g55389、LOC_Os04g55510、LOC_Os08g07740和LOC_Os08g01670共6个基因在双亲间的表达量差异显著, 其中LOC_Os03g50310在Nekken2中的表达量比HZ高3.6倍。对双亲间候选基因LOC_Os03g50310进行测序分析, 发现该基因在5′UTR、CDS区及3′UTR存在4处差异, 其中CDS区的单核苷酸多态性(SNP)差异引发单个氨基酸改变。研究通过挖掘水稻抽穗期QTL位点为进一步克隆水稻抽穗期相关基因和品种选育提供了新线索。
利用高密度Bin遗传图谱定位水稻抽穗期QTL
DOI:10.3724/SP.J.1006.2023.12089
[本文引用: 1]

挖掘新的控制水稻抽穗期相关位点和候选基因, 对抽穗期的遗传机制研究和品种改良具有重要的意义。利用抽穗期存在明显差异的粳稻TD70和籼稻Kasalath杂交衍生的包含186个家系的重组自交系群体, 构建了基于深度重测序的高密度Bin遗传图谱, 图谱共包含12,328个Bin标记。RIL群体及亲本2018年和2021年正季种植于江苏省南京市江苏省农业科学院。以家系从播种到抽穗所经历的天数作为抽穗期表型值, 使用IciMapping软件3.4版的完备区间作图法, 对控制水稻抽穗期的QTL进行鉴定。2年共检测到15个抽穗期的QTL, 分布在3号、6号、7号、8号、10号和12号染色体上, LOD值为2.58~10.68, 其中7个QTL和已知抽穗期QTL的位置存在重叠或者部分重叠。共有4个QTL在2年均检测到, 表现出较强的稳定性。对2年重复鉴定到的4个QTL区间进行基因功能注释和亲本间序列分析, 共发现7个注释有功能, 且在2个亲本间编码区存在非同义突变的基因。根据候选基因SNP的类型对RIL群体家系进行基因等位型分类和效应分析, 发现4个基因其不同等位型的RIL家系在抽穗期上存在显著或者极显著差异, 推测可能为候选基因, 可用于后续水稻抽穗期的分子机制研究。
The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors
DOI:10.1016/0092-8674(95)90288-0
PMID:7697715
[本文引用: 1]

The vegetative and reproductive (flowering) phases of Arabidopsis development are clearly separated. The onset of flowering is promoted by long photoperiods, but the constans (co) mutant flowers later than wild type under these conditions. The CO gene was isolated, and two zinc fingers that show a similar spacing of cysteines, but little direct homology, to members of the GATA1 family were identified in the amino acid sequence. co mutations were shown to affect amino acids that are conserved in both fingers. Some transgenic plants containing extra copies of CO flowered earlier than wild type, suggesting that CO activity is limiting on flowering time. Double mutants were constructed containing co and mutations affecting gibberellic acid responses, meristem identity, or phytochrome function, and their phenotypes suggested a model for the role of CO in promoting flowering.
Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS
Photoperiod and temperature synergistically regulate heading date and regional adaptation in rice
Hd1 function conversion in regulating heading is dependent on gene combinations of Ghd7, Ghd8, and Ghd7.1 under long-day conditions in rice
水稻光周期调控开花的研究进展
DOI:10.16819/j.1001-7216.2021.0514
[本文引用: 1]

水稻抽穗期作为重要的农艺性状,由自身遗传因素和环境因素共同决定,对品种生态适应区域和产量因子均有较大影响。过去的二十年里,从叶片的日长识别到茎尖分生组织的成花激活,水稻光周期诱导抽穗开花分子调控机理已取得较大进展,分离并克隆大量与成花相关的调控基因,并整合到光周期调控分子网络中。当植物处于有利条件时,该网络激活成花调控基因,促进成花素表达,将成花素运输至顶端分生组织,从而驱动分生组织细胞发育,最终成花。本文以拟南芥为对照参考,对水稻光周期调控网络及由低纬度地区向高纬度地区扩展遗传变异进行讨论,以期为生态型品种培育和光周期调控成花分子机理研究提供参考。
Days to heading,controlled by the heading date genes, Hd1 and DTH8, limits rice yield-related traits in Hokkaido, Japan
DOI:10.1270/jsbbs.19151
PMID:32714049
[本文引用: 2]

A key aspect of rice breeding programs is the optimization of days to heading (DTH) for maximizing grain productivity in cultivation areas. Here, the effects of genotypes for heading date on yield-related traits in rice (culm and panicle length (CL and PL), panicle number (PN), and total number of seeds) were investigated. () and () are the main controllers of the variation in heading date in the rice population of Hokkaido, Japan. Thus, an F population (n = 192) derived from a cross between Kitaibuki () and Akage () was developed. Significant differences in DTH were found among all combinations. Each genotype for heading date showed variations in the yield-related traits without a significant difference. However, DTH exhibited high positive coefficient values (more than 0.709) with the yield-related traits except for PN, which had a negative coefficient value of -0.431. A later heading date resulted in a longer growth duration and a higher yield with a combination of longer PL and CL and lower PN. These results suggest that DTH limits the yield-related traits rather than the genotype for heading date.Copyright © 2020 by JAPANESE SOCIETY OF BREEDING.
Ehd1,a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1
Homodimerization of Ehd1 is required to induce flowering in rice
Dual function of clock component OsLHY sets critical day length for photoperiodic flowering in rice
Effect of the relative length of day and night and other factors of the environment on growth and reproduction in plants
Photoperiodic flowering: time measurement mechanisms in leaves
DOI:10.1146/annurev-arplant-043014-115555
PMID:25534513
[本文引用: 2]

Many plants use information about changing day length (photoperiod) to align their flowering time with seasonal changes to increase reproductive success. A mechanism for photoperiodic time measurement is present in leaves, and the day-length-specific induction of the FLOWERING LOCUS T (FT) gene, which encodes florigen, is a major final output of the pathway. Here, we summarize the current understanding of the molecular mechanisms by which photoperiodic information is perceived in order to trigger FT expression in Arabidopsis as well as in the primary cereals wheat, barley, and rice. In these plants, the differences in photoperiod are measured by interactions between circadian-clock-regulated components, such as CONSTANS (CO), and light signaling. The interactions happen under certain day-length conditions, as previously predicted by the external coincidence model. In these plants, the coincidence mechanisms are governed by multilayered regulation with numerous conserved as well as unique regulatory components, highlighting the breadth of photoperiodic regulation across plant species.
Circadian clock and photoperiodic flowering in Arabidopsis:CONSTANS is a hub for signal integration
Environmental signal-dependent regulation of flowering time in rice
OsPhyA modulates rice flowering time mainly through OsGI under short days and Ghd7 under long days in the absence of phytochrome B
A base substitution in OsphyC disturbs its interaction with OsphyB and affects flowering time and chlorophyll synthesis in rice
Molecular dissection of the roles of phytochrome in photoperiodic flowering in rice
DOI:10.1104/pp.111.181792
PMID:21880933
[本文引用: 3]

Phytochromes mediate the photoperiodic control of flowering in rice (Oryza sativa), a short-day plant. Recent molecular genetics studies have revealed a genetic network that enables the critical daylength response of florigen gene expression. Analyses using a rice phytochrome chromophore-deficient mutant, photoperiod sensitivity5, have so far revealed that within this network, phytochromes are required for expression of Grain number, plant height and heading date7 (Ghd7), a floral repressor gene in rice. There are three phytochrome genes in rice, but the roles of each phytochrome family member in daylength response have not previously been defined. Here, we revealed multiple action points for each phytochrome in the critical daylength response of florigen expression by using single and double phytochrome mutant lines of rice. Our results show that either phyA alone or a genetic combination of phyB and phyC can induce Ghd7 mRNA, whereas phyB alone causes some reduction in levels of Ghd7 mRNA. Moreover, phyB and phyA can affect Ghd7 activity and Early heading date1 (a floral inducer) activity in the network, respectively. Therefore, each phytochrome gene of rice has distinct roles, and all of the phytochrome actions coordinately control the critical daylength response of florigen expression in rice.
Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions
DOI:10.1093/pcp/pcf156
PMID:12407188
[本文引用: 1]

Heading date 3a (Hd3a) has been detected as a heading-date-related quantitative trait locus in a cross between rice cultivars Nipponbare and Kasalath. A previous study revealed that the Kasalath allele of Hd3a promotes heading under short-day (SD) conditions. High-resolution linkage mapping located the Hd3a locus in a approximately 20-kb genomic region. In this region, we found a candidate gene that shows high similarity to the FLOWERING LOCUS T (FT) gene, which promotes flowering in Arabidopsis: Introduction of the gene caused an early-heading phenotype in rice. The transcript levels of Hd3a were increased under SD conditions. The rice Heading date 1 (Hd1) gene, a homolog of CONSTANS (CO), has been shown to promote heading under SD conditions. By expression analysis, we showed that the amount of Hd3a mRNA is up-regulated by Hd1 under SD conditions, suggesting that Hd3a promotes heading under the control of Hd1. These results indicate that Hd3a encodes a protein closely related to Arabidopsis FT and that the function and regulatory relationship with Hd1 and CO, respectively, of Hd3a and FT are conserved between rice (an SD plant) and Arabidopsis (a long-day plant).
A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice
DOI:10.1242/dev.040170
PMID:19762423
[本文引用: 1]

Although some genes that encode sensory or regulatory elements for photoperiodic flowering are conserved between the long-day (LD) plant Arabidopsis thaliana and the short-day (SD) plant rice, the gene networks that control rice flowering, and particularly flowering under LD conditions, are not well understood. We show here that RICE FLOWERING LOCUS T 1 (RFT1), the closest homolog to Heading date 3a (Hd3a), is a major floral activator under LD conditions. An RFT1:GFP fusion protein localized in the shoot apical meristem (SAM) under LD conditions, suggesting that RFT1 is a florigen gene in rice. Furthermore, mutants in OsMADS50, a rice ortholog of Arabidopsis SUPPRESOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) did not flower up to 300 days after sowing under LD conditions, indicating that OsMADS50, which acts upstream of RFT1, promotes flowering under LD conditions. We propose that both positive (OsMADS50 and Ehd1) and negative (Hd1, phyB and Ghd7) regulators of RFT1 form a gene network that regulates LD flowering in rice. Among these regulators, Ehd1, a rice-specific floral inducer, integrates multiple pathways to regulate RFT1, leading to flowering under appropriate photoperiod conditions. A rice ortholog of Arabidopsis APETALA1, OsMADS14, was expressed in the floral meristem in wild-type but not in RFT1 RNAi plants, suggesting that OsMADS14 is activated by RFT1 protein in the SAM after the transition to flowering. We have thus exposed a network of genes that regulate LD flowering in rice.
OsGI controls flowering time by modulating rhythmic flowering time regulators preferentially under short day in rice
OsLHY is involved in regulating flowering through the Hd1- and Ehd1- mediated pathways in rice (Oryza sativa L.)
The clock component OsLUX regulates rice heading through recruiting OsELF3-1 and OsELF4s to repress Hd1 and Ghd7
Ehd2, a rice ortholog of the maize INDETERMINATE1 gene, promotes flowering by up-regulating Ehd1
Ehd3, encoding a plant homeodomain finger-containing protein, is a critical promoter of rice flowering
DOI:10.1111/j.1365-313X.2011.04517.x
PMID:21284756
[本文引用: 2]

Oryza sativa (rice) flowers in response to photoperiod, and is a facultative short-day (SD) plant. Under SD conditions, flowering is promoted through the activation of FT-like genes (rice florigens) by Heading date 1 (Hd1, a rice CONSTANS homolog) and Early heading date 1 (Ehd1, with no ortholog in the Arabidopsis genome). On the other hand, under long-day (LD) conditions, flowering is delayed by the repressive function of Hd1 on FT-like genes and by downregulation of Ehd1 by the flowering repressor Ghd7 - a unique pathway in rice. We report here that an early heading date 3 (ehd3) mutant flowered later than wild-type plants, particularly under LD conditions, regardless of the Hd1-deficient background. Map-based cloning revealed that Ehd3 encodes a nuclear protein that contains a putative transcriptional regulator with two plant homeodomain (PHD) finger motifs. To identify the role of Ehd3 within the gene regulatory network for rice flowering, we compared the transcript levels of genes related to rice flowering in wild-type plants and ehd3 mutants. Increased transcription of Ghd7 under LD conditions and reduced transcription of downstream Ehd1 and FT-like genes in the ehd3 mutants suggested that Ehd3 normally functions as an LD downregulator of Ghd7 in floral induction. Furthermore, Ehd3 ghd7 plants flowered earlier and show higher Ehd1 transcript levels than ehd3 ghd7 plants, suggesting a Ghd7-independent role of Ehd3 in the upregulation of Ehd1. Our results demonstrate that the PHD-finger gene Ehd3 acts as a promoter in the unique genetic pathway responsible for photoperiodic flowering in rice.© 2011 The Authors. The Plant Journal © 2011 Blackwell Publishing Ltd.
Ehd4 Encodes a Novel and Oryza-genus-specific regulator of photoperiodic flowering in rice
Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice
Ef7 encodes an ELF3-like protein and promotes rice flowering by negatively regulating the floral repressor gene Ghd7 under both short- and long-day conditions
Natural variation in Hd17, a homolog of Arabidopsis ELF3 that is involved in rice photoperiodic flowering
Fine-tuning Flowering Time via genome editing of upstream open reading frames of Heading Date 2 in rice
DOI:10.1186/s12284-021-00504-w
PMID:34189630
[本文引用: 2]

Flowering time of rice (Oryza sativa L.) is among the most important agronomic traits for region adaptation and grain yield. In the process of rice breeding, efficient and slightly modulating the flowering time of an elite cultivar would be more popular with breeder. Hence, we are interested in slightly increasing the expression of flowering repressors by CRISPR/Cas9 genome editing system. It was predicated there were three uORFs in 5' leader sequence of Hd2. In this study, through editing Hd2 uORFs, we got four homozygous mutant lines. Phenotypic analysis showed that the hd2 urf edited lines flowered later by 4.6-11.2 days relative to wild type SJ2. Supporting the later flowering phenotype, the expression of Ehd1, Hd3a, and RFT1 is significantly decreased in hd2 urf than that in wild type. Moreover, we found that the transcription level of Hd2 is not affected, whereas the Hd2 protein level was increased in hd2 urf compared with wild type, which indicated that Hd2 uORFs indeed affect the translation of a downstream Hd2 pORF. In summary, we developed a efficient approach for delaying rice heading date based on editing uORF region of flowering repressor, which is time and labor saving compared to traditional breeding. In future, uORF of other flowering time related genes, including flowering promoter and flowering repressor genes, can also be used as targets to fine-tune the flowering time of varieties.
DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously
OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a
Heading date gene, dth3 controlled late flowering in O. Glaberrima Steud. by down- regulating Ehd1
DOI:10.1007/s00299-011-1129-4
PMID:21830130
[本文引用: 1]

Heading date in rice is an important agronomic trait controlled by several genes. In this study, flowering time of variety Dianjingyou 1 (DJY1) was earlier than a near-isogenic line (named NIL) carried chromosome segment from African rice on chromosome 3S, when grown in both long-day (LD) and short-day (SD) conditions. By analyzing a large F2 population from NIL × DJY1, the locus DTH3 (QTL for days to heading on chromosome 3) controlling early heading date in DJY1 was fine mapped to a 64-kb segment which contained only one annotated gene, a MIKC-type MADS-box protein. We detected a 6-bp deletion and a single base substitution in the C-domain by sequencing DTH3 in DJY1 compared with dth3 in NIL, and overexpression of DTH3 caused early flowering in callus. Quantitative real-time PCR revealed that the transcript level of dth3 in NIL was lower than that DTH3 in DJY1 in both LD and SD conditions. The Early heading date 1 (Ehd1) which promotes the RFT1, was up-regulated by DTH3 in both LD and SD conditions. Based on Indel and dCAPs marker analysis, the dth3 allele was only present in African rice accessions. A phylogenetic analysis based on microsatellite genotyping suggested that African rice had a close genetic relationship to O. rufipogon and O. latifolia, and was similar to japonica cultivars. DTH3 affected flowering time and had no significant effect on the main agronomic traits.
OsBBX14 delays heading date by repressing florigen gene expression under long and short-day conditions in rice
DOI:10.1016/j.plantsci.2016.02.017
PMID:27095397
[本文引用: 3]

B-box (BBX) proteins are zinc finger proteins containing B-box domains, which have roles in Arabidopsis growth and development. However, little is known concerning rice BBXs. Herein, we identified a rice BBX protein, Oryza sativa BBX14 (OsBBX14). OsBBX14 is highly expressed in flag leaf blades. OsBBX14 expression shows a diurnal rhythm under photoperiodic conditions and subsequent continuous white light. OsBBX14 is located in the nucleus and has transcriptional activation potential. OsBBX14-overexpression (OsBBX14-OX) lines exhibited delayed heading date under long-day (LD) and short-day (SD) conditions, whereas RNAi lines of OsBBX14 lines had similar heading dates to the WT. The florigen genes, Hd3a and RFT1, were downregulated in the OsBBX14-OX lines under LD and SD conditions. Under LD conditions, Hd1 was expressed higher in the OsBBX14-OX lines than in the wild type (WT), and the rhythmic expression of circadian clock genes, OsLHY and OsPRR1, was changed in OsBBX14-OX lines. Thus, OsBBX14 acts as a floral repressor by promoting Hd1 expression under LD conditions, probably because of crosstalk with the circadian clock. Under SD conditions, Ehd1 expression was reduced in OsBBX14-OX lines, but Hd1 and circadian clock gene expressions were unaffected, indicating that OsBBX14 acts as a repressor of Ehd1. Our findings suggested that OsBBX14 regulates heading date differently under LD and SD conditions. Copyright © 2016 Elsevier Ireland Ltd. All rights reserved.
An operational definition of epigenetics
Autophagy targets Hd1 for vacuolar degradation to regulate rice flowering
Hd1, Ghd7, and DTH8 synergistically determine the rice heading date and yield-related agronomic traits
OsFLZ2 interacts with OsMADS51 to fine-tune rice flowering time
Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice
Proceedings of the National Academy of Sciences of the United States of America,
Phytochromes are the sole photoreceptors for perceiving red/far-red light in rice
Proceedings of the National Academy of Sciences of the United States of America,
Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice
Post-transcriptional regulation of Ghd 7 protein stability by phytochrome and OsGI in photoperiodic control of flowering in rice
Time to flower: interplay between photoperiod and the circadian clock
DOI:10.1093/jxb/eru441
PMID:25371508
[本文引用: 1]

Plants precisely time the onset of flowering to ensure reproductive success. A major factor in seasonal control of flowering time is the photoperiod. The length of the daily light period is measured by the circadian clock in leaves, and a signal is conveyed to the shoot apex to initiate floral transition accordingly. In the last two decades, the molecular players in the photoperiodic pathway have been identified in Arabidopsis thaliana. Moreover, the intricate connections between the circadian clockwork and components of the photoperiodic pathway have been unravelled. In particular, the molecular basis of time-of-day-dependent sensitivity to floral stimuli, as predicted by Bünning and Pittendrigh, has been elucidated. This review covers recent insights into the molecular mechanisms underlying clock regulation of photoperiodic responses and the integration of the photoperiodic pathway into the flowering time network in Arabidopsis. Furthermore, examples of conservation and divergence in photoperiodic flower induction in other plant species are discussed. © The Author 2014. Published by Oxford University Press on behalf of the Society for Experimental Biology. All rights reserved. For permissions, please email: journals.permissions@oup.com.
The transcriptional repressor OsPRR73 links circadian clock and photoperiod pathway to control heading date in rice
Genetic relationship between phytochromes and OsELF3-1 reveals the mode of regulation for the suppression of phytochrome signaling in rice
Induction of flowering by DNA demethylation in Perilla frutescens and Silene armeria: Heritability of 5-azacytidine-induced effects and alteration of the DNA methylation state by photoperiodic conditions
Knockdown of SAMS genes encoding S-adenosyl-L-methionine synthetases causes methylation alterations of DNAs and histones and leads to late flowering in rice
组蛋白修饰在水稻中的研究进展
DOI:10.13304/j.nykjdb.2021.0497
[本文引用: 1]

作为表观遗传学研究的重要内容,组蛋白修饰在维持真核生物基因组稳定性、基因表达调控和染色质结构等方面发挥重要作用。水稻是重要的粮食作物,也是科学研究的模式植物。近年来研究发现,组蛋白修饰参与了水稻生长发育、胁迫应答、产量以及品质形成等重要生物学性状的调控。因此,明确组蛋白修饰在水稻中的遗传和调控机制对于水稻遗传改良具有重要意义。对组蛋白修饰的作用机制以及在水稻中的研究进展进行了综述,并进一步展望了水稻中组蛋白修饰的研究前景,以期为水稻育种提供参考。
SET DOMAIN GROUP701 encodes a H3K4-methytransferase and regulates multiple key processes of rice plant development
The rice enhancer of zeste [E(z)] genes SDG711 and SDG718 are respectively involved in long day and short day signaling to mediate the accurate photoperiod control of flowering time
LC2 and OsVIL 2 promote rice flowering by photoperoid-induced epigenetic silencing of OsLF
DOI:10.1093/mp/sss096
PMID:22973062
[本文引用: 1]

Proper flowering time is essential for plant reproduction. Winter annual Arabidopsis thaliana needs vernalization before flowering, during which AtVILs (VIN3 and VRN5, components of PRC2 complex) mediate the H3K27 tri-methylation at the FLC locus (a floral repressor) to repress the FLC expression and hence to induce flowering. However, how VILs (VIL, VERNALIZATION INSENSITIVE 3-LIKE) function in rice is unknown. Here we demonstrated that rice LC2 (OsVIL3) and OsVIL2 (two OsVILs, possible components of PRC2 complex) promote rice flowering. Our results showed that expressions of LC2 and OsVIL2 are induced by SD (short-day) conditions and both lc2 mutant and OsVIL2-RNAi lines display delayed heading date, consistent with the reduced expression levels of Hd1 and Hd3a. Interestingly, LC2 binds to the promoter region of a floral repressor OsLF and represses the OsLF expression via H3K27 tri-methylation modification. In addition, OsLF directly regulates the Hd1 expression through binding to Hd1 promoter. These results first demonstrated that the putative PRC2 in rice is involved in photoperiod flowering regulation, which is different from that of Arabidopsis, and revealed that LC2 binds the promoter region of target gene, presenting a possible mechanism of the recruitment process of PRC2 complex to its target genes. The studies provide informative clues on the epigenetic control of rice flowering.
OsVIL1 controls flowering time in rice by suppressing OsLF under short days and by inducing Ghd7 under long days
The WD40 domain- containing protein Ehd5 positively regulates flowering in rice (Oryza sativa)
The RING-finger ubiquitin ligase HAF 1 mediates heading date 1 degradation during photoperiodic flowering in rice
SPIN1, a K homology domain protein negatively regulated and ubiquitinated by the E 3 ubiquitin ligase SPL11, is involved in flowering time control in rice
OsHUB2 inhibits function of OsTrx1 in heading date in rice
Interaction between temperature and photoperiod in regulation of flowering time in rice
High ambient temperatures inhibit Ghd7-Mediated flowering repression in rice
Dynamic effects of interacting genes underlying rice flowering-time phenotypic plasticity and global adaptation
DOI:10.1101/gr.255703.119
PMID:32299830
[本文引用: 1]

The phenotypic variation of living organisms is shaped by genetics, environment, and their interaction. Understanding phenotypic plasticity under natural conditions is hindered by the apparently complex environment and the interacting genes and pathways. Herein, we report findings from the dissection of rice flowering-time plasticity in a genetic mapping population grown in natural long-day field environments. Genetic loci harboring four genes originally discovered for their photoperiodic effects (,,, and ) were found to differentially respond to temperature at the early growth stage to jointly determine flowering time. The effects of these plasticity genes were revealed with multiple reaction norms along the temperature gradient. By coupling genomic selection and the environmental index, accurate performance predictions were obtained. Next, we examined the allelic variation in the four flowering-time genes across the diverse accessions from the 3000 Rice Genomes Project and constructed haplotypes at both individual-gene and multigene levels. The geographic distribution of haplotypes revealed their preferential adaptation to different temperature zones. Regions with lower temperatures were dominated by haplotypes sensitive to temperature changes, whereas the equatorial region had a majority of haplotypes that are less responsive to temperature. By integrating knowledge from genomics, gene cloning and functional characterization, and environment quantification, we propose a conceptual model with multiple levels of reaction norms to help bridge the gaps among individual gene discovery, field-level phenotypic plasticity, and genomic diversity and adaptation.© 2020 Guo et al.; Published by Cold Spring Harbor Laboratory Press.
The importance of being on time: regulatory networks controlling photoperiodic flowering in cereals
DOI:10.3389/fpls.2017.00665
PMID:28491078
[本文引用: 1]

Flowering is the result of the coordination between genetic information and environmental cues. Gene regulatory networks have evolved in plants in order to measure diurnal and seasonal variation of day length (or photoperiod), thus aligning the reproductive phase with the most favorable season of the year. The capacity of plants to discriminate distinct photoperiods classifies them into long and short day species, depending on the conditions that induce flowering. Plants of tropical origin and adapted to short day lengths include rice, maize, and sorghum, whereas wheat and barley were originally domesticated in the Fertile Crescent and are considered long day species. In these and other crops, day length measurement mechanisms have been artificially modified during domestication and breeding to adapt plants to novel areas, to the extent that a wide diversity of responses exists within any given species. Notwithstanding the ample natural and artificial variation of day length responses, some of the basic molecular elements governing photoperiodic flowering are widely conserved. However, as our understanding of the underlying mechanisms improves, it becomes evident that specific regulators exist in many lineages that are not shared by others, while apparently conserved components can be recruited to novel functions during evolution.
DELLA-dependent and -independent gibberellin signaling
Rice early flowering1, a CKI, phosphorylates DELLA protein SLR1 to negatively regulate gibberellin signalling
DOI:10.1038/emboj.2010.75
PMID:20400938
[本文引用: 1]

The plant hormone gibberellin (GA) is crucial for multiple aspects of plant growth and development. To study the relevant regulatory mechanisms, we isolated a rice mutant earlier flowering1, el1, which is deficient in a casein kinase I that has critical roles in both plants and animals. el1 had an enhanced GA response, consistent with the suppression of EL1 expression by exogenous GA(3). Biochemical characterization showed that EL1 specifically phosphorylates the rice DELLA protein SLR1, proving a direct evidence for SLR1 phosphorylation. Overexpression of SLR1 in wild-type plants caused a severe dwarf phenotype, which was significantly suppressed by EL1 deficiency, indicating the negative effect of SLR1 on GA signalling requires the EL1 function. Further studies showed that the phosphorylation of SLR1 is important for maintaining its activity and stability, and mutation of the candidate phosphorylation site of SLR1 results in the altered GA signalling. This study shows EL1 a novel and key regulator of the GA response and provided important clues on casein kinase I activities in GA signalling and plant development.
RID1 sets rice heading date by balancing its binding with SLR1 and SDG722
DOI:10.1111/jipb.13196
[本文引用: 1]

Rice (<i>Oryza sativa</i>) is a major crop that feeds billions of people, and its yield is strongly influenced by flowering time (heading date). Loss of <i>RICE INDETERMINATE1</i> (<i>RID1</i>) function causes plants not to flower; thus, <i>RID1</i> is considered a master switch among flowering-related genes. However, it remains unclear whether other proteins function together with RID1 to regulate rice floral transition. Here, we revealed that the chromatin accessibility and H3K9ac, H3K4me3, and H3K36me3 levels at <i>Heading date 3a</i> (<i>Hd3a</i>) and <i>RICE FLOWERING LOCUS T1</i> (<i>RFT1</i>) loci were significantly reduced in <i>rid1</i> mutants. Notably, RID1 interacted with SET DOMAIN GROUP PROTEIN 722 (SDG722), a methyltransferase. We determined that SDG722 affects the global level of H3K4me2/3 and H3K36me2/3, and promotes flowering primarily through the <i>Early heading date1</i>-<i>Hd3a</i>/<i>RFT1</i> pathway. We further established that rice DELLA protein SLENDER RICE1 (SLR1) interacted with RID1 to inhibit its transactivation activity, that <i>SLR1</i> suppresses rice flowering, and that messenger RNA and protein levels of SLR1 gradually decrease with plant growth. Furthermore, SLR1 competed with SDG722 for interaction with RID1. Overall, our results establish that interplay between RID1, SLR1, and SDG722 feeds into rice flowering-time control.
Exogenous abscisic acid represses rice flowering via SAPK8-ABF1-Ehd1/Ehd2 pathway
Drought stress in rice: morpho-physiological and molecular responses and marker- assisted breeding
Hd3a, RFT1 and Ehd1 integrate photoperiodic and drought stress signals to delay the floral transition in rice
Integrative regulation of drought escape through ABA-dependent and -independent pathways in rice
A drought-inducible transcription factor delays reproductive timing in rice
DOI:10.1104/pp.16.01691
PMID:26945049
[本文引用: 1]

The molecular mechanisms underlying photoperiod or temperature control of flowering time have been recently elucidated, but how plants regulate flowering time in response to other external factors, such as water availability, remains poorly understood. Using a large-scale Hybrid Transcription Factor approach, we identified a bZIP transcriptional factor, O. sativa ABA responsive element binding factor 1 (OsABF1), which acts as a suppressor of floral transition in a photoperiod-independent manner. Simultaneous knockdown of both OsABF1 and its closest homologous gene, OsbZIP40, in rice (Oryza sativa) by RNA interference results in a significantly earlier flowering phenotype. Molecular and genetic analyses demonstrate that a drought regime enhances expression of the OsABF1 gene, which indirectly suppresses expression of the Early heading date 1 (Ehd1) gene that encodes a key activator of rice flowering. Furthermore, we identified a drought-inducible gene named OsWRKY104 that is under the direct regulation of OsABF1 Overexpression of OsWRKY104 can suppress Ehd1 expression and confers a later flowering phenotype in rice. Together, these findings reveal a novel pathway by which rice modulates heading date in response to the change of ambient water availability.© 2016 American Society of Plant Biologists. All Rights Reserved.
中国水稻种植区划
根据各地生态环境、社会经济条件和水稻种植特点,将中国稻区划分为6个稻作区(一级区)和16个稻作亚区(二级区)。对各个稻作区和亚区的基本情况、自然生态条件、稻田种植制度、品种类型、主要病虫害等作了评述,並提出水稻生产发展方向、途径和措施。
Association of functional nucleotide polymorphisms at DTH2 with the northward expansion of rice cultivation in Asia
Proceedings of the National Academy of Sciences of the United States of America,
Genetic and molecular dissection of flowering time control in rice//Rice Genomics, Genetics and Breeding
Heading date 1 (Hd1), on ortholog of Arabidopsis CONSTANS, is a possible target of human selection during domestication to diversify flowering times of cultivated rice
OsPRR37 and Ghd7 are the major genes for general combining ability of DTH, PH and SPP in rice
Genetic interactions among Ghd7, Ghd8, OsPRR37 and Hd1 contribute to large variation in heading date in rice
DOI:10.1186/s12284-019-0314-x
PMID:31309345
[本文引用: 1]

Heading date is crucial for rice reproduction and geographic expansion. Many heading date genes are sensitive to photoperiod and jointly regulate flowering time in rice. However, it is not clear how these genes coordinate rice heading.Here, we performed a genetic interaction analysis among four major rice heading date genes Ghd7, Ghd8, OsPRR37/Ghd7.1 (hereafter PRR37) and Hd1 in the near-isogenic background under both natural long-day (NLD) and natural short-day (NSD) conditions. The 4-gene segregating population exhibited a large heading date variation with more than 95 days under NLD and 42 days under NSD conditions. Tetragenic, trigenic and digenic interactions among these four genes were observed under both conditions but more significant under NLD conditions. In the functional Hd1 backgrounds, the strongest digenic interaction was Ghd7 by Ghd8 under NLD but was Ghd7 by PRR37 under NSD conditions. Interestingly, PRR37 acted as a flowering suppressor under NLD conditions, while it functioned alternatively as an activator or a suppressor under NSD conditions depending on the status of the other three genes. Based on the performances of 16 homozygous four-gene combinations, a positive correlation between heading date and spikelets per panicle (SPP) was found under NSD conditions, but changed to a negative correlation when heading date was over 90 days under NLD conditions.These results demonstrate the importance of genetic interactions in the rice flowering regulatory network and will help breeders to select favorable combinations to maximize rice yield potential for different ecological areas.
Enhancing yield and improving grain quality in japonica rice: Targeted EHD1 editing via CRISPR-Cas9 in low-latitude adaptation
DOI:10.3390/cimb46040233
PMID:38666963
[本文引用: 1]

The " to " initiative in China focuses on adapting rice varieties from the northeast to the unique photoperiod and temperature conditions of lower latitudes. While breeders can select varieties for their adaptability, the sensitivity to light and temperature often complicates and prolongs the process. Addressing the challenge of cultivating high-yield, superior-quality rice over expanded latitudinal ranges swiftly, in the face of these sensitivities, is critical. Our approach harnesses the CRISPR-Cas9 technology to edit the gene in the premium northeastern cultivars Jiyuanxiang 1 and Yinongxiang 12, which are distinguished by their exceptional grain quality-increased head rice rates, gel consistency, and reduced chalkiness and amylose content. Field trials showed that these new mutants not only surpass the wild types in yield when grown at low latitudes but also retain the desirable traits of their progenitors. Additionally, we found that disabling boosts the activity of and, postponing flowering by approximately one month in the mutants. This research presents a viable strategy for the accelerated breeding of elite northeastern rice by integrating genomic insights with gene-editing techniques suitable for low-latitude cultivation.