作物杂志,2016, 第2期: 14–20 doi: 10.16035/j.issn.1001-7283.2016.02.003
植物Dof转录因子的结构特点及功能研究进展
张雪1,2,尹悦佳2,范贝1,李慧杰1,费小钰1,崔喜艳1
- 1 吉林农业大学生命科学学院/作物营养代谢与调控实验室,130118,吉林长春
2 吉林省农业科学院农业生物技术研究所,130124,吉林长春
Advances on the Structural Characteristics and Function of Dof Transcription Factors in Plant
Zhang Xue1,2,Yin Yuejia2,Fan Bei1,Li Huijie1,Fei Xiaoyu1,Cui Xiyan1
- 1 School of Life Sciences,Jilin Agricultural University/Laboratory of Crop Nutrition Metabolism and Regulation,Changchun 130118,Jilin,China
2 Institute of Agricultural Biotechnology,Jilin Academy of Agricultural Sciences,Changchun 130124,Jilin,China
摘要:
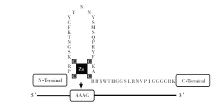
Dof(DNA binding with one finger)蛋白是植物中特有的转录因子,其N-末端具含保守锌指结构的Dof结构域,能够识别AAAG序列;Dof转录因子能够发挥多种功能,主要是由于其具有多变的C-末端,它是Dof蛋白的特异转录调控结构域。在植物中,Dof转录因子的功能主要与维管发育、叶片极性、保卫细胞特异基因的调控、碳氮代谢、种子发育和萌发、光响应及光周期调控的开花和花粉发育、次生代谢物合成、防御反应、生长素响应等生理过程有关。将这些进程归纳为组织分化、种子发育和代谢调节三个方面,通过对植物Dof转录因子的结构特点及功能进行分析,为Dof转录因子的深入研究提供参考。
| [1] | Yanagisawa S, Izui K . Molecular cloning of two DNA-binding proteins of maize that are structurally different but interact with the same sequence motif. Journal of Biological Chemistry, 1993,268(21):16028-16036. |
| [2] |
Gupta S, Malviya N, Kushwaha H , et al. Insights into structural and functional diversity of Dof(DNA binding with one finger) transcription factor. Planta, 2015,241:549-562.
doi: 10.1007/s00425-014-2239-3 |
| [3] |
Moreno-Risueno M A, Martnez M, Vicente-Carbajosa J , et al. The family of DOF transcription factors:from green unicellular algae to vascular plants. Molecular Genetics and Genomics, 2007,277(4):379-390.
doi: 10.1007/s00438-006-0186-9 |
| [4] |
Lijavetzky D, Carbonero P, Vicente-Carbajosa J . Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evolutionary Biology, 2003,3:17.
doi: 10.1186/1471-2148-3-17 |
| [5] |
Moreno-Risueno M A, Diaz I, Carrillo L , et al. The HvDOF19 transcription factor mediates the abscisic acid-dependent repression of hydrolase genes in germinating barley aleurone. The Plant Journal, 2007,51(3):352-365.
doi: 10.1111/j.1365-313X.2007.03146.x |
| [6] | Guo Y, Qiu L J . Genome-wide analysis of the Dof transcription factor gene family reveals soybean specific duplicable and functional characteristics. PLoS ONE, 2013,8(9):1-35. |
| [7] |
Yang X, Tuskan G A, Cheng M Z . Divergence of the Dof gene families in poplar,Arabidopsis,and rice suggests multiple modes of gene evolution after duplication. Plant Physiology, 2006,142(3):820-830.
doi: 10.1104/pp.106.083642 |
| [8] | Shaw L M ,McIntyre C L ,Gresshoff P M,et al.Members of the Dof transcription factor family in Triticum aestivum are associated with light mediated gene regulation. Functional & Integrative Genomics, 2009,9(4):485-498. |
| [9] |
Kushwaha H, Gupta S, Singh V K , et al. Genome wide identification of Dof transcription factor gene family in sorghum and its comparative phylogenetic analysis with rice and Arabidopsis. Molecular Biology Reports, 2011,38(8):5037-5053.
doi: 10.1007/s11033-010-0650-9 |
| [10] |
Jiang Y, Zeng B, Zhao H , et al. Genome-wide transcription factor gene prediction and their expressional tissue specificities in maize. Journal of Integrative Plant Biology, 2012,54(9):616-630.
doi: 10.1111/j.1744-7909.2012.01149.x |
| [11] |
Hernando-Amado S, González-Calle V, Carbonero P , et al. The family of DOF transcription factors in Brachypodium distachyon:phylogenetic comparison with rice and barley DOFs and expression profiling. BMC Plant Biology, 2012,12:202.
doi: 10.1186/1471-2229-12-202 |
| [12] |
Cai X, Zhang Y, Zhang C , et al. Genome-wide analysis of plant specific Dof transcription factor family in tomato. Journal of Integrative Plant Biology, 2013,55(6):552-566.
doi: 10.1111/jipb.v55.6 |
| [13] |
Gupta S, Kushwaha H ,Singh V K et al.Genome wide in silico characterization of Dof transcription factor gene family of sugarcane and its comparative phylogenetic analysis with Arabidopsis,rice and sorghum. Sugar Tech, 2014,16(4):372-384.
doi: 10.1007/s12355-013-0288-8 |
| [14] | Malviya N, Gupta S, Singh V K , et al. Genome wide in silico characterization of Dof gene families of pigeonpea [Cajanus cajan(L) Millisp.]. Molecular Biology Reports, 2014,42(2):535-552. |
| [15] |
Yanagisawa S, Schmidt R J . Diversity and similarity among recognition sequences of Dof transcription factors. Plant Journal, 1999,17(2):209-214.
doi: 10.1046/j.1365-313X.1999.00363.x |
| [16] |
Diaz I, Vicente-Carbajosa J, Abraham Z , et al. The GAMYB protein from barley interacts with the Dof transcription factor BPBF and activates endosperm specific genes during seed development. Plant Journal, 2002,29(4):453-464.
doi: 10.1046/j.0960-7412.2001.01230.x |
| [17] |
Yanagisawa S . Dof domain proteins:plant-specific transcription factors associated with diverse phenomena unique to plants. Plant and Cell Physiology, 2004,45(4):386-391.
doi: 10.1093/pcp/pch055 |
| [18] |
Umemura Y, Ishiduka T, Yamamoto R , et al. The Dof domain,a zinc finger DNA-binding domain conserved only in higher plants,truly functions as a Cys2/Cys2 Zn finger domain. Plant Journal, 2004,37(5):741-749.
doi: 10.1111/tpj.2004.37.issue-5 |
| [19] |
Kisu Y, Ono T, Shimofurutani N , et al. Characterization and expression of a new class of zinc finger protein that binds to silencer region of ascorbate oxidase gene. Plant and Cell Physiology, 1998,39(10):1054-1064.
doi: 10.1093/oxfordjournals.pcp.a029302 |
| [20] |
Cominelli E, Galbiati M, Albertini A , et al. DOF-binding sites additively contribute to guard cell-specificity of AtMYB60 promoter. BMC Plant Biology, 2011,11(1):1-13.
doi: 10.1186/1471-2229-11-1 |
| [21] |
Yang J, Yang M F, Zhang W P , et al. A putative flowering-time-related Dof transcription factor gene,JcDof3,is controlled by the circadian clock in Jatropha curcas. Plant Science, 2011,181(6):667-674.
doi: 10.1016/j.plantsci.2011.05.003 |
| [22] |
Chen X, Wang D, Liu C , et al. Maize transcription factor Zmdof1 involves in the regulation of Zm401 gene. Plant Growth Regulation, 2012,66(3):271-284.
doi: 10.1007/s10725-011-9651-5 |
| [23] |
Yanagisawa S . The Dof family of plant transcription factors. Trends in Plant Science, 2002,7(12):555-560.
doi: 10.1016/S1360-1385(02)02362-2 |
| [24] |
Yanagisawa S . The transcriptional activation domain of the plant-specific Dof1 factor functions in plant,animal,and yeast cells. Plant Cell Physiology, 2001,42(8):813-822.
doi: 10.1093/pcp/pce105 |
| [25] |
Krebs J, Mueller-Roeber B, Ruzicic S . A novel bipartite nuclear localization signal with an atypically long linker in Dof transcription factors. Journal of Plant Physiology, 2010,167(7):583-586.
doi: 10.1016/j.jplph.2009.11.016 |
| [26] | Kushwaha H, Gupta N, Singh V K , et al. In silico analysis of PCR amplified Dof (DNA binding with one finger)transcription factor domain and cloned genes from cereals and millets. Bioinformatics, 2008,9(1):130-143. |
| [27] |
Noguero M, Atif R M, Ochatt S , et al. The role of the DNA-binding One Zinc Finger(Dof) transcription factor family in plants. Plant Science, 2013,209:32-45.
doi: 10.1016/j.plantsci.2013.03.016 |
| [28] |
Kim H S, Kim S J, Abbasi N , et al. The Dof transcription factor Dof5.1 influences leaf axial patterning by promoting revolute transcription in Arabidopsis. Plant Journal, 2010,64(3):524-535.
doi: 10.1111/tpj.2010.64.issue-3 |
| [29] |
Konishi M, Yanagisawa S . Sequential activation of two Dof transcription factor gene promoters during vascular development in Arabidopsis thaliana. Plant Physiology and Biochemistry, 2007,45(8):623-629.
doi: 10.1016/j.plaphy.2007.05.001 |
| [30] |
De Paolis A, Sabatini S, De Pascalis L , et al. A rolB regulatory factor belongs to a new class of single zinc finger plant proteins. Plant Journal, 1996,10(2):215-223.
doi: 10.1046/j.1365-313X.1996.10020215.x |
| [31] |
Baumann K, De Paolis A, Costantino P , et al. The DNA binding site of the Dof protein NtBBF1 is essential for tissue-specific and auxin-regulated expression of the rolB oncogene in plants. Plant Cell, 1999,11(3):323-334.
doi: 10.1105/tpc.11.3.323 |
| [32] |
Kang H G, Singh K B . Characterization of salicylic acid-responsive,Arabidopsis Dof domain proteins:overexpression of OBP3 leads to growth defects. Plant Journal, 2000,21(4):329-339.
doi: 10.1046/j.1365-313x.2000.00678.x |
| [33] | Yu J, Shi G, Yu D . Constitutive overexpression of GmDof17-1,a putative Dof transcription factor from soybean causing growth inhibition in tobacco. Scientia Agricola, 2013,71(1):44-51. |
| [34] | Plesch G, Ehrhardt T, Mueller-Roeber B . Involvement of TAAAG elements suggests a role for Dof transcription factors in guard cell-specific gene expression. Plant Journal, 2001,28(4):455-464. |
| [35] |
Negi J, Moriwaki K, Konishi M , et al. A Dof transcription factor,SCAP1,is essential for the development of functional stomata in Arabidopsis. Current Biology, 2013,23(6):479-484.
doi: 10.1016/j.cub.2013.02.001 |
| [36] |
Skirycz A, Radziejwoski A, Busch W , et al. The Dof transcription factor OBP1 is involved in cell cycle regulation in Arabidopsis thaliana. Plant Journal, 2008,56(7):779-792.
doi: 10.1111/tpj.2008.56.issue-5 |
| [37] |
Song Y H, Smith R W, To B J , et al. FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science, 2012,336(6084):1045-1049.
doi: 10.1126/science.1219644 |
| [38] |
Corrales A R, Sergio G N, Laura C , et al. Characterization of tomato cycling Dof factors reveals conserved and new functions in the control of flowering time and abiotic stress responses. Journal of Experimental Botany, 2014,65(4):995-1012.
doi: 10.1093/jxb/ert451 |
| [39] |
Ahmad M, Rim Y, Chen H , et al. Functional characterization of Arabidopsis Dof transcription factor AtDof4.1. Russian Journal Plant Physiology, 2013,60(1):116-123.
doi: 10.1134/S1021443712060027 |
| [40] |
Wei P C, Tan F, Gao X Q , et al. Overexpression of AtDof4.7,an Arabidopsis Dof family transcription factor,induces floral organ abscission deficiency in Arabidopsis. Plant Physiology, 2010,153(3):1031-1045.
doi: 10.1104/pp.110.153247 |
| [41] |
Marzábal P, Gas E, Fontanet P , et al. The maize Dof protein PBF activates transcription of gamma-zein during maize seed development. Plant Molecular Biology, 2008,67(5):441-454.
doi: 10.1007/s11103-008-9325-5 |
| [42] |
Kawakatsu T, Takaiwa F . Differences in transcriptional regulatory mechanisms functioning for free lysine content and seed storage protein accumulation in rice grain. Plant and Cell Physiology, 2010,51(12):1964-1974.
doi: 10.1093/pcp/pcq164 |
| [43] |
Wang H W, Zhang B, Hao Y J , et al. The soybean Dof-type transcription factor genes,GmDof4 and GmDof11,enhance lipid content in the seeds of transgenic Arabidopsis plants. Plant Journal, 2007,52(4):716-729.
doi: 10.1111/j.1365-313X.2007.03268.x |
| [44] | 王志坤 , Arun S, 常健敏, 等.转GmDof11基因高油转基因大豆的鉴定及主要农艺性状调查.作物杂志, 2014(2):39-42. |
| [45] | Zhang Y . Functional analysis of Dof transcription factors controlling heading date and PPDK gene expression in rice. Leiden:Leiden University, 2015. |
| [46] | Gabriele S, Rizza A, Martone J , et al. The Dof protein DAG1 mediates PIL5 activity on seed germination by negatively regulating GA biosynthetic gene AtGA3ox1. Plant Journal, 2010,61(2):312-323. |
| [47] |
Rizza A, Boccaccini A, Lopez-Vidriero I , et al. Inactivation of the ELIP1 and ELIP2 genes affects Arabidopsis seed germination. New Phytologist, 2011,190(4):896-905.
doi: 10.1111/nph.2011.190.issue-4 |
| [48] |
Isabel-LaMoneda I, Diaz I, Martinez M , et al. SAD:a new Dof protein from barley that activates transcription of a cathepsin B-like thiol protease gene in the aleurone of germinating seeds. Plant Journal, 2003,33(2):329-340.
doi: 10.1046/j.1365-313X.2003.01628.x |
| [49] |
Zou X, Neuman D, Shen Q J . Interactions of two transcriptional repressors and two transcriptional activators in modulating gibberellin signaling in aleurone cells. Plant Physiology, 2008,148(1):176-186.
doi: 10.1104/pp.108.123653 |
| [50] | Washio K . Identification of Dof proteins with implication in the gibberellin-regulated expression of a peptidase gene following the germination of rice grains. Biochimica et Biophysica Acta Journal, 2001,1520(1):54-62. |
| [51] |
Rueda-Romero P, Barrero-Sicilia C, Gomez-Cadenas A , et al. Arabidopsis thaliana Dof6 negatively affects germination in non-after-ripened seeds and interacts with TCP14. Journal Experimental Botany, 2012,63(5):1937-1949.
doi: 10.1093/jxb/err388 |
| [52] |
Yanagisawa S, Sheen J . Involvement of maize Dof zinc finger proteins in tissue-specific and light-regulated gene expression. Plant Cell, 1998,10(1):75-89.
doi: 10.1105/tpc.10.1.75 |
| [53] |
Kurai T, Wakayama M, Abiko T , et al. Introduction of the ZmDof1 gene into rice enhances carbon and nitrogen assimilation under low-nitrogen conditions. Plant Biotechnology Journal, 2011,9(8):826-837.
doi: 10.1111/j.1467-7652.2011.00592.x |
| [54] |
Cavalar M, Phlippen Y, Kreuzaler F , et al. A drastic reduction in Dof1 transcript levels does not affect C4-specific gene expression in maize. Journal of Plant Physiology, 2007,164(12):1665-1674.
doi: 10.1016/j.jplph.2006.09.008 |
| [55] |
Tanaka M, Takahata Y, Nakayama H , et al. Altered carbohydrate metabolism in the storage roots of sweet potato plants overexpressing the SRF1 gene,which encodes a Dof zinc finger transcription factor. Planta, 2009,230(4):737-746.
doi: 10.1007/s00425-009-0979-2 |
| [56] |
Santos L A , Souza S R D,Fernandes M S.OsDof25 expression alters carbon and nitrogen metabolism in Arabidopsis under high N-supply. Plant Biotechnology Reports, 2012,6(4):327-337.
doi: 10.1007/s11816-012-0227-2 |
| [57] |
Skirycz A, Jozefczuk S, Stobiecki M , et al. Transcription factor AtDof4;2 affects phenylpropanoid metabolism in Arabidopsis thaliana. New Phytologist, 2007,175(3):425-438.
doi: 10.1111/nph.2007.175.issue-3 |
| [58] |
Park D H, Lim P O, Kim J S , et al. The Arabidopsis COG1 gene encodes a Dof domain transcription factor and negatively regulates phytochrome signaling. Plant Journal, 2003,34(2):161-171.
doi: 10.1046/j.1365-313X.2003.01710.x |
| [59] |
Papi M, Sabatini S, Altamura M L , et al. Inactivation of the phloem-specific Dof zinc finger gene DAG1 affects response to light and integrity of the testa of Arabidopsis seeds. Plant Physiology, 2002,128(2):411-417.
doi: 10.1104/pp.010488 |
| [60] |
Iwamoto M, Higo K, Takano M . Circadian clock-and phytochrome-regulated Dof-like gene,Rdd1,is associated with grain size in rice. Plant Cell and Environment, 2009,32(5):592-603.
doi: 10.1111/pce.2009.32.issue-5 |
| [61] | Yang J, Yang M F, Wang D , et al. JcDof1,a Dof transcription factor gene,is associated with the light-mediated circadian clock in Jatropha curcas. Physiology Plant, 2010,139(3):324-334. |
| [62] |
Rueda-Lopez M, Crespillo R, Canovas F M , et al. Differential regulation of two glutamine synthetase genes by a single Dof transcription factor. Plant Journal, 2008,56(1):73-85.
doi: 10.1111/tpj.2008.56.issue-1 |
| [63] | 胡娟, 王丽鸳, 韦康 , 等. 茶树CsCDF1基因克隆及表达分析. 茶叶科学, 2015,35(5):501-511. |
| [64] |
Skirycz A, Reichelt M, Burow M , et al. Dof transcription factor AtDof1.1(OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in Arabidopsis. Plant Journal, 2006,47(1):10-24.
doi: 10.1111/tpj.2006.47.issue-1 |
| [65] |
Nakano T, Suzuki K, Ohtsuki N , et al. Identification of genes of the plant specific transcription factor families cooperatively regulated by ethylene and jasmonate in Arabidopsis thaliana. Journal Plant Research, 2006,119(4):407-413.
doi: 10.1007/s10265-006-0287-x |
| [1] | 王丰丰 王 萍 陈 利 代彩玲 李书颉 张贺平. 基于植物学性状分析籽用南瓜的形态多样性[J]. 作物杂志, 2018, (5): 77–84 |
| [2] | 曹玉巧,聂庆凯,高云,许自成黄五星,. 植物中镉及其螯合物相关转运蛋白研究进展[J]. 作物杂志, 2018, (3): 15–24 |
| [3] | 熊伟姣,王亚伦,姚绍嫦,潘春柳,肖冬,王爱勤,何龙飞. MicroRNA在高等植物逆境响应中的作用机制研究进展[J]. 作物杂志, 2018, (1): 1–8 |
| [4] | 赵振杰,梁太波,陈千思,胡利伟,张艳玲,尹启生. 碳纳米材料对植物生长发育的调节作用[J]. 作物杂志, 2017, (2): 7–13 |
| [5] | 段俊枝,李莹,赵明忠,李清州,张莉,魏小春,任银铃. NAC转录因子在植物抗非生物胁迫基因工程中的应用进展[J]. 作物杂志, 2017, (2): 14–22 |
| [6] | 罗昊文,孔雷蕾,钟卓君,谢萝雅,赵旭生,林子秋,唐湘如. 淹水胁迫对水稻玉香油占秧苗生长和生理特性的影响[J]. 作物杂志, 2017, (1): 135–139 |
| [7] | 高志勇,谢恒星,王志平,刘史力. 植物突变体库的作用及构建研究进展[J]. 作物杂志, 2016, (6): 16–19 |
| [8] | 余永廷,高春生,李智敏,曾粮斌,陈佳,朱爱国,朱涛涛,孙凯,严准. Phytocystatin在植物抗病虫中的作用及表达调控[J]. 作物杂志, 2016, (6): 9–15 |
| [9] | 柏祥,塔莉,赵美微,刘传才,魏国印,崔力拓,曾广娟,秦津. 外来入侵植物反枝苋的最新研究进展[J]. 作物杂志, 2016, (4): 7–14 |
| [10] | 韩赞平,陈彦惠,郭书磊,祖小峰,王顺喜,赵西拥. 玉米抗逆基因ZmqLTG3-1的克隆及功能分析[J]. 作物杂志, 2016, (4): 47–55 |
| [11] | 左应梅,杨维泽,杨天梅,杨美权,许宗亮,杨绍兵,张金渝. 干旱胁迫下4种人参属植物抗性生理指标的比较[J]. 作物杂志, 2016, (3): 84–88 |
| [12] | 袁月,郑有飞,赵辉,储仲芳,黄积庆. 臭氧与其他环境因子及其复合作用对植物的影响[J]. 作物杂志, 2016, (1): 16–22 |
| [13] | 李洋, 李春雷, 路明, 等. 植物激素、维生素引发老化玉米种子的研究[J]. 作物杂志, 2015, (6): 150–154 |
| [14] | 赵翠荣, 刘莹, 唐清, 等. Dicamba和2,4-D处理对小麦与玉米杂交得胚率的影响[J]. 作物杂志, 2015, (5): 46–49 |
| [15] | 李丹丹, 闫丽, 常健敏, 等. 大豆GmWRI1基因在糖、植物激素及盐胁迫下的表达分析[J]. 作物杂志, 2015, (4): 41–46 |
|