作物杂志,2019, 第6期: 168–176 doi: 10.16035/j.issn.1001-7283.2019.06.027
基于qPCR和LAMP技术的马铃薯晚疫病菌快速检测方法
祝菊澧1,2,3,梁静思1,2,3,张佩1,王伟伟1,2,3,林桐司骐1,谢欣娱1,苏瑞1,唐唯1,2,3
- 1云南师范大学生命科学学院,650500,云南昆明
2云南师范大学马铃薯科学研究院,650500,云南昆明
3云南省马铃薯生物学重点实验室,650500,云南昆明
A Rapid Detection Method for Potato Late Blight Caused by Phytophthora infestans Based on qPCR and LAMP Assays
Zhu Juli1,2,3,Liang Jingsi1,2,3,Zhang Pei1,Wang Weiwei1,2,3,Lin Tongsiqi1,Xie Xinyu1,Su Rui1,Tang Wei1,2,3
- 1School of Life Science, Yunnan Normal University, Kunming 650500, Yunnan, China
2Joint Academy of Potato Science, Yunnan Normal University, Kunming 650500, Yunnan, China
3Key Laboratory of Potato Biology of Yunnan Province, Kunming 650500, Yunnan, China
摘要:
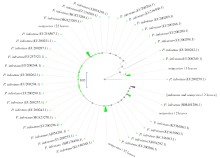
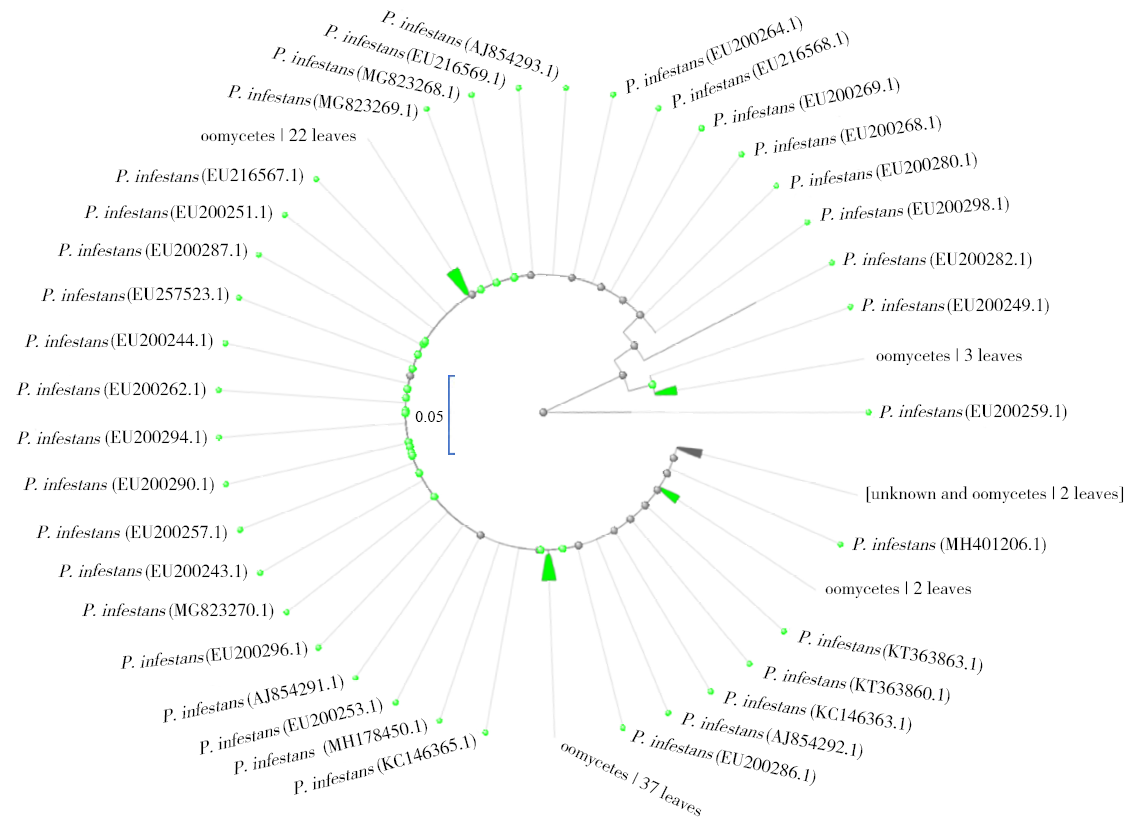
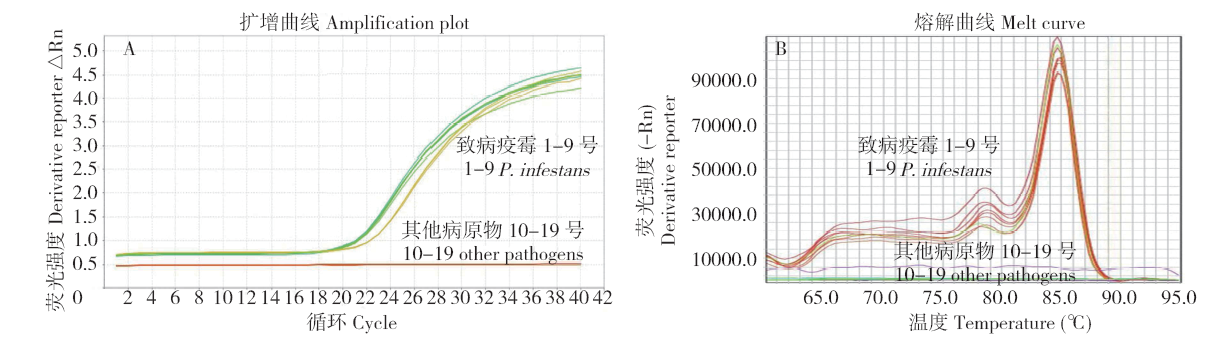
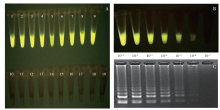
马铃薯晚疫病菌(Phytophthora infestans)能侵染多种茄科植物,它引起的马铃薯晚疫病,是马铃薯生产中的第一大病害。为了开发能在田间快速检测马铃薯晚疫病病原的方法,利用P. infestans T30-4基因组测序数据的contig 1.18131,设计qPCR和LAMP引物,优化扩增条件后得到引物的特异性和灵敏度,最后通过检测田间收获薯块,比较形态学传统方法、qPCR及LAMP的差异。特异性检测结果发现,qPCR和LAMP仅在含有P. infestans DNA模板的体系有阳性扩增,在寄主和其他微生物DNA中均无扩增;在优化的条件下,qPCR和LAMP的检测下限可达1×10 -6ng/μL,在有寄主和其他微生物DNA存在的条件下,引物的灵敏度没有显著差异。利用两种快速方法对在大理、丽江及昆明3个地区田间收获薯块上检测发现,qPCR和LAMP方法得到的检出率差异极为不显著(P=0.420),两种快速检测方法和形态学鉴定方法检出率差异极显著(P=0.009)。在大理、丽江及昆明3个地区的薯块中,两种分子检测方法检出率均比形态学方法高。其中,qPCR检测方法比形态学方法分别提高了12.00%、2.00%、8.70%;LAMP检测方法比形态学方法分别提高了11.30%、2.00%、8.70%。
| [1] |
Fry W E . Phytophthora infestans:the plant (and R gene) destroyer. Molecular Plant Pathology, 2008,9(3):385-402.
doi: 10.1111/j.1364-3703.2007.00465.x pmid: 18705878 |
| [2] |
Fry W E, Birch P R J, Judelson H S ,et al. Five reasons to consider Phytophthora infestans a reemerging pathogen. Phytopathology, 2015,105(7):966-981.
doi: 10.1094/PHYTO-01-15-0005-FI pmid: 25760519 |
| [3] |
Yuen J E, Andersson B . What is the evidence for sexual reproduction of Phytophthora infestans in Europe?. Plant Pathology, 2013,62(3):485-491.
doi: 10.1111/j.1365-3059.2012.02685.x |
| [4] |
Dennisa J, Thomas F C . Latent infection of potato seed tubers by Phytophthora infestans during long-term cold storage. Plant Disease, 2009,93(9):940-946.
doi: 10.1094/PDIS-93-9-0940 pmid: 30754539 |
| [5] |
Small I M, Joseph L, Fry W E . Development and implementation of the BlightPro decision support system for potato and tomato late blight management. Computers and Electronics in Agriculture, 2015,115:57-65.
doi: 10.1016/j.compag.2015.05.010 |
| [6] |
Fall M L, Tremblay D M, Gobeil-Richard M , et al. Infection efficiency of four Phytophthora infestans clonal lineages and DNA-based quantification of sporangia. PLoS ONE, 2015,10(8):e0136312.
doi: 10.1371/journal.pone.0136312 pmid: 26301826 |
| [7] |
Trout C L, Ristaino J B, Madritch M , et al. Rapid detection of Phytophthora infestans in late blight-infected potato and tomato using PCR. Plant Disease, 1997,81(9):1042-1048.
doi: 10.1094/PDIS.1997.81.9.1042 pmid: 30861957 |
| [8] |
Ristaino J B, Madritch M, Trout C L , et al. PCR amplification of ribosomal DNA for species identification in the plant pathogen genus Phytophthora. Applied and Environmental Microbiology, 1998,64(3):948-954.
pmid: 9501434 |
| [9] |
Kong P, Hong C X, Richardson P A , et al. Single-strand-conformation polymorphism of ribosomal DNA for rapid species differentiation in genus Phytophthora. Fungal Genetics and Biology, 2003,39(3):238-249.
doi: 10.1016/S1087-1845(03)00052-5 |
| [10] |
Tooley P W, Bunyard B A, Carras M M , et al. Development of PCR primers from internal transcribed spacer region 2 for detection of Phytophthora species infecting potatoes. Applied and Environmental Microbiology, 1997,63(4):1467-1475.
pmid: 9097445 |
| [11] |
Hussain T, Sharma S, Singh B P , et al. Detection of latent infection of Phytophthora infestans in potato seed tubers. Potato Journal, 2013,40(2):142-148.
doi: 10.1007/BF02849142 |
| [12] |
Judelson H S, Tooley P W . Enhanced polymerase chain reaction methods for detecting and quantifying Phytophthora infestans in plants. Phytopathology, 2000,90(10):1112-1119.
doi: 10.1094/PHYTO.2000.90.10.1112 pmid: 18944474 |
| [13] | Gómez-Alpizar L, Carbone I, Ristaino J B . An Andean origin of Phytophthora infestans inferred from mitochondrial and nuclear gene genealogies. Proceedings of the National Academy of Sciences of the United States of America, 2007,104(9):3306-3311. |
| [14] |
Llorente B, Bravoalmonacid F, Cvitanich C , et al. A quantitative real-time PCR method for in planta monitoring of Phytophthora infestans growth. Letters in Applied Microbiology, 2010,51(6):603-610.
doi: 10.1111/j.1472-765X.2010.02942.x pmid: 21039667 |
| [15] |
Mehran K, Benjin L, Yue J , et al. Evaluation of different PCR-based assays and LAMP method for rapid detection of Phytophthora infestans by targeting the Ypt1 gene. Frontiers in Microbiology, 2017,8:1920.
doi: 10.3389/fmicb.2017.01920 pmid: 29051751 |
| [16] |
Hussain S, Lees A K, Duncan J M , et al. Development of a species-specific and sensitive detection assay for Phytophthora infestans and its application for monitoring of inoculum in tubers and soil. Plant Pathology, 2005,54(3):373-382.
doi: 10.1111/ppa.2005.54.issue-3 |
| [17] |
Ammour M S, Bilodeau G J, Tremblay D M , et al. Development of real-time isothermal amplification assays for on-site detection of Phytophthora infestans in potato leaves. Plant Disease, 2017,101(7):1269-1277.
doi: 10.1094/PDIS-12-16-1780-RE pmid: 30682973 |
| [18] |
Notomi T, Okayama H, Masubuchi H , et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Research, 2000,28(12):e63.
doi: 10.1093/nar/28.12.e63 pmid: 10871386 |
| [19] |
Nagamine K, Watanabe K, Ohtsuka K , et al. Loop-mediated isothermal amplification reaction using a nondenatured template. Clinical Chemistry, 2001,47(9):1742-1743.
pmid: 11514425 |
| [20] |
Martin F N, Abad Z G, Balci Y , et al. Identification and detection of Phytophthora:reviewing our progress,identifying our needs. Plant Disease, 2012,96(8):1080-1103.
doi: 10.1094/PDIS-12-11-1036-FE pmid: 30727075 |
| [21] |
Tomlinson J A, Barker I, Boonham N . Faster,simpler,more-specific methods for improved molecular detection of Phytophthora ramorum in the field. Applied and Environmental Microbiology, 2007,73(12):4040-4047.
doi: 10.1128/AEM.00161-07 pmid: 17449689 |
| [22] |
Hansen Z R, Knaus B J, Tabima J F , et al. Loop-mediated isothermal amplification for detection of the tomato and potato late blight pathogen,Phytophthora infestans. Journal of Applied Microbiology, 2016,120(4):1010-1020.
doi: 10.1111/jam.13079 pmid: 26820117 |
| [23] |
Chen Y, Roxby R . Characterization of a Phytophthora infestans gene involved in vesicle transport. Gene, 1996,181(1/2):89-94.
doi: 10.1016/s0378-1119(96)00469-6 pmid: 8973313 |
| [24] |
Caten C E, Jinks J L . Spontaneous variability of single isolates of Phytophthora infestans. I. cultural variation. Canadian Journal of Botany, 1968,46:329-348.
doi: 10.1139/b68-055 |
| [25] |
Álvarez I, Wendel J F . Ribosomal ITS sequences and plant phylogenetic inference. Molecular Phylogenetics and Evolution, 2003,29(3):417-434.
doi: 10.1016/s1055-7903(03)00208-2 pmid: 14615184 |
| [26] |
Lane D J . Nucleic acid techniques in bacterial systematics. New York:John Wiley and Sons, 1991.
doi: 10.3760/cma.j.issn.0253-9624.2019.11.011 pmid: 31683400 |
| [27] | 魏景超 . 真菌鉴定手册. 上海: 上海科学技术出版社, 1979. |
| [28] | Boone D R, Castenholz R W . Bergey′s manual of systematic bacteriology. New York:Springer, 2001. |
| [29] |
Kroon L P, Brouwer H, de Cock A W ,et al. The genus Phytophthora anno 2012. Phytopathology, 2012,102(4):348.
doi: 10.1094/PHYTO-01-11-0025 |
| [30] |
Blancomeneses M, Ristaino J B . Detection and quantification of Peronospora tabacina using a real-time polymerase chain reaction assay. Plant Disease, 2011,95(6):673-682.
doi: 10.1094/PDIS-05-10-0333 |
| [31] |
Diguta C F, Rousseaux S, Weidmann S , et al. Development of a qPCR assay for specific quantification of Botrytis cinereal on grapes. Fems Microbiology Letters, 2010,313(1):81-87.
doi: 10.1111/j.1574-6968.2010.02127.x pmid: 20946385 |
| [32] | Spring O, Marco T, Wolf S , et al. PCR-based detection of sunflower white blister rust (Pustula helianthicola C. Rost and Thines) in soil samples and asymptomatic host tissue. Netherlands Journal of Plant Pathology, 2011,131(3):519-527. |
| [33] |
Eshraghi L, Anderson J, Aryamanesh N , et al. Phosphite primed defence responses and enhanced expression of defence genes in Arabidopsis thaliana infected with Phytophthora cinnamomic. Plant Pathology, 2011,60(6):1086-1095.
doi: 10.1111/j.1365-3059.2011.02471.x |
| [34] |
Lees A K, Sullivan L, Lynott J S , et al. Development of a quantitative real-time PCR assay for Phytophthora infestans and its applicability to leaf,tuber and soil samples. Plant Pathology, 2012,61(5):867-876.
doi: 10.1111/j.1365-3059.2011.02574.x |
| [1] | 周弋力,张亚玲,赵宏森,靳学慧. 黑龙江省主栽水稻品种抗稻瘟病基因的分子检测与分析[J]. 作物杂志, 2019, (3): 172–177 |
| [2] | 李丽娜,金龙国,谢传晓,刘昌林. 转基因玉米和转基因大豆盲样检测方法[J]. 作物杂志, 2017, (6): 37–44 |
| [3] | 赵欣欣,宋丽荣,权薇竹,曲兴然,王奇. 高温高湿老化处理对不同豆类种子活力及生长能力的影响[J]. 作物杂志, 2017, (5): 168–172 |
| [4] | 卢艳丽,周洪友,张笑宇. 马铃薯茎尖脱毒方法优化及病毒检测[J]. 作物杂志, 2017, (1): 161–167 |
| [5] | 郭翠,张维,余桂容,周正富,李亮,冯帅,陈明,王劲. 转G2-EPSPS基因玉米D-3侧翼序列分析与转化体特异性检测方法[J]. 作物杂志, 2016, (1): 69–75 |
| [6] | 谢传晓, 李新海, 张世煌. 农产品未准人转基因成分“低水平混杂”的概念、成因及对策[J]. 作物杂志, 2015, (3): 1–4 |
| [7] | 李兴欣, 孟义江, 罗婉娇, 等. 基于绿色荧光标记的甘草遗传转化体系的建立[J]. 作物杂志, 2014, (4): 52–58 |
| [8] | 蒋运斌, 马逾英, 杨枝中, 等. 川白芷种子水溶性内源性萌发抑制物质的初步研究[J]. 作物杂志, 2014, (3): 95–100 |
| [9] | 刘建伟, 陈晓峰, 刘广富, 郭宗端, 李新柱, 胡兆平, 张亮. 大豆CYP78A5基因组织特异性启动子的克隆及表达分析[J]. 作物杂志, 2014, (1): 54–58 |
| [10] | 崔少彬. 农杆菌介导OD}EB2B基因转化中薯3号的研究[J]. 作物杂志, 2013, (6): 46–49 |
| [11] | 邹月利, 陶波. 除草剂氯嘧磺隆在培养基中的提取和检测方法研究[J]. 作物杂志, 2012, (6): 48–51 |
| [12] | 王淑芳, 马桂珍, 暴增海, 李世东, 陈月, 钱媛媛, 尹璐. 辣椒疫霉菌(Phytophthora capsici)实时荧光定量检测方法的研究[J]. 作物杂志, 2012, (5): 30–35 |
| [13] | 滕卫丽, 韩英鹏, 赵桂云, 等. 大豆叶黄素的研究进展[J]. 作物杂志, 2012, (1): 9–12 |
| [14] | 李婷婷, 韩岚岚, 赵奎军, 等. 圆盘分割法在转基因大豆抗虫鉴定上的应用[J]. 作物杂志, 2011, (4): 20–22 |
| [15] | 白月, 才华, 栾凤侠, 等. 多重PCR结合DHPLC方法检测番茄中转基因成分[J]. 作物杂志, 2011, (2): 28–31 |
|