作物杂志,2021, 第1期: 16–25 doi: 10.16035/j.issn.1001-7283.2021.01.003
玉米ZmGS5基因克隆、分子特性分析及对拟南芥的遗传转化
刘晓丽1,2( ), 韩利涛1, 魏楠1, 申飞1, 蔡一林2(
), 韩利涛1, 魏楠1, 申飞1, 蔡一林2( )
)
- 1河南农业职业学院食品工程学院,451450,河南郑州
2西南大学玉米研究所,400715,重庆
Cloning, Molecular Characteristics Analysis and Genetic Transformation of Arabidopsis thaliana of ZmGS5 Gene in Maize
Liu Xiaoli1,2( ), Han Litao1, Wei Nan1, Shen Fei1, Cai Yilin2(
), Han Litao1, Wei Nan1, Shen Fei1, Cai Yilin2( )
)
- 1Department of Food Engineering, Henan Vocational College of Agriculture, Zhengzhou 451450, Henan, China
2Maize Research Institute, Southwest University, Chongqing 400715, China
摘要:
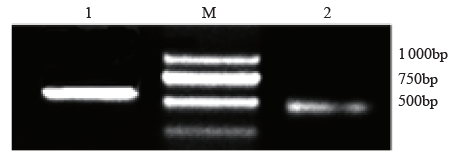
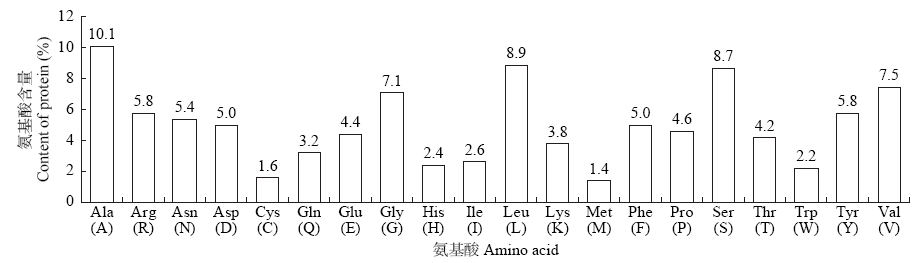
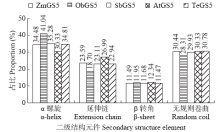
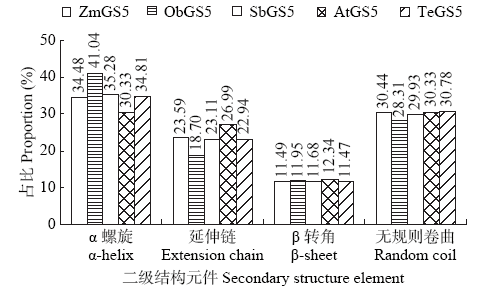
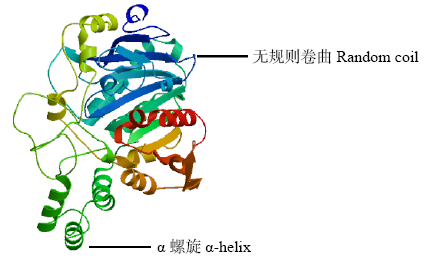
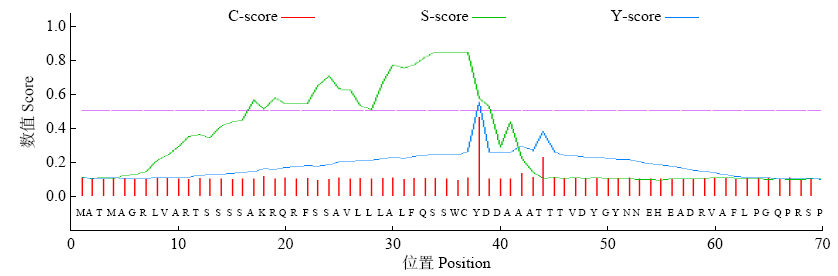
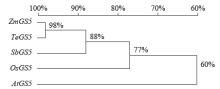
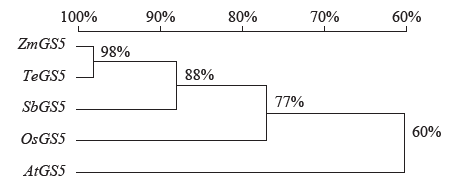
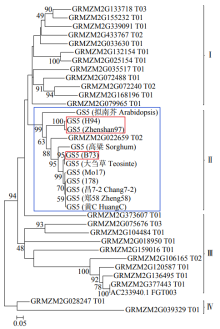
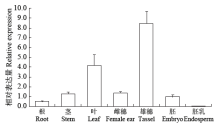
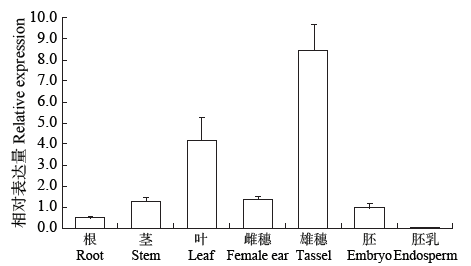
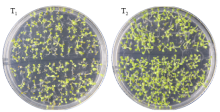
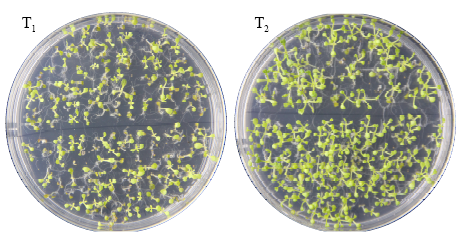
高产是玉米育种的重要目标,籽粒大小是决定籽粒产量的重要因子。基于水稻中控制籽粒大小的OsGS5基因编码序列,利用同源克隆方法,获得了玉米ZmGS5基因,其cDNA全长为1 695bp,开放阅读框1 491bp,编码496个氨基酸,含有Peptidase-S10结构域、1个信号肽和1个丝氨酸羧肽酶(serine carboxypeptidases,SCP)类蛋白所特有的催化活性中心,这些都与SCP家族结构特点相符,也与OsGS5相关研究结果相同;磷酸化位点分析结果表明,ZmGS5含有丝氨酸、苏氨酸和酪氨酸等蛋白激酶识别位点。qRT-PCR分析显示ZmGS5在雄穗和叶片中的表达量较高,而在胚及胚乳中的表达量相对较低。除此之外,采用农杆菌介导法,建立了拟南芥的遗传转化体系,得到纯合转基因株系,T3代转基因拟南芥种子千粒重为0.0169g,较野生型千粒重(0.0139g)高。
| [1] | 李向拓, 毛建昌, 吴权明. 分子标记在玉米育种中的应用. 玉米科学, 2004,43(2):26-29. |
| [2] | 贾波, 管飞翔, 谢庆春, 等. 玉米产量性状QTL定位分析. 西南农业学报, 2013,26(1):22-25. |
| [3] | 黄荣荣, 周子键, 陈甲法, 等. 玉米缺陷性籽粒突变体的遗传分析及突变基因dek1-T7的定位. 分子植物育种, 2012,10(2):163-168. |
| [4] |
Li X, Chen G H, Zhang W Y, et al. Genome-wide transcriptional analysis of maize endosperm in response to ae wx double mutations. Journal of Genetics and Genomics, 2010,37(11):749-762.
doi: 10.1016/S1673-8527(09)60092-8 pmid: 21115169 |
| [5] |
Yi G, Lauter A M, Scott M P, et al. The thick aleurone 1 mutant defines a negative regulation of maize aleurone cell fate that functions downstream of defective kernell. Plant Physiology, 2011,156:1826-1836.
doi: 10.1104/pp.111.177725 pmid: 21617032 |
| [6] | 宋同明, 陆效武. 对一个玉米双重标记新突变基因(os)的染色体定位和初步遗传研究. 遗传学报, 1993(5):432-438. |
| [7] | Lid S E, Gruis D, Jung R, et al. The defective kernel 1 (dek1) gene require for aleurone cell development in the endosperm of maize grains encodes a membrane protein of the calpain gene superfamily. Proceedings of the National Academy of Sciences of the United States of America, 2002,8(99):5460-5465. |
| [8] |
Jahnke S, Scholten S. Epigenetic resetting of a gene imprinted in plant embryos. Current Biology, 2009,19:1677-1681.
doi: 10.1016/j.cub.2009.08.053 pmid: 19781944 |
| [9] |
Li Y, Fan C, Xing Y, et al. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nature Genetics, 2011,43(12):1266-1269.
doi: 10.1038/ng.977 pmid: 22019783 |
| [10] |
Kellogg E A. Evolutionary history of the grasses. Plant Physiology, 2001,125(3):1198-1205.
pmid: 11244101 |
| [11] | 王建兵, 汤华, 黄益勤, 等. 玉米和水稻重要性状QTL的比较研究. 遗传学报, 2004,31(12):1401-1407. |
| [12] |
Raith M R, Kelty C A, Griffith J F, et al. Comparison of PCR and quantitative real-time PCR methods for the characterization of ruminant and cattle fecal pollution sources. Water Research, 2013,47(18):6921-6928.
doi: 10.1016/j.watres.2013.03.061 pmid: 23871256 |
| [13] | 刘晓丽, 魏楠. 拟南芥的遗传转化. 河南农业, 2019(27):45-46. |
| [14] | Xu C J, Liu Y, Li Y B, et al. Differential expression of GS5 regulates grain size in rice. Journal of Experimental Botany, 2015,9(66):2611-2623. |
| [15] | 胡甘雨, 司风铃, 车燕飞, 等. 葱蝇过氧化氢酶基因的克隆及生物信息学分析. 西南大学学报(自然科学版), 2014,36(2):32-40. |
| [16] |
Mahoney J A, Ntolosi B, DaSilva R P, et al. Cloning and characterization of CPVL,a novel serine carboxypeptidase,from human macrophages. Genomics, 2001,72:243-251.
doi: 10.1006/geno.2000.6484 pmid: 11401439 |
| [17] | Shirley A M, Chapple C. Biochemical characterization of sinapoyglucose choline sinapolytransferase,a serine carboxypeptidase-like protein that functions as an acyltransferase in plant secondary metabolism. Journal Biological Chemistry, 2003,278(22):19870-19877. |
| [18] |
Cercós M, Urbez C, Carbonell J. A serine carboxypeptidase gene (PsCP),expressed in early steps of reproductive and vegetative development in Pisum sativum,is induced by gibberellins. Plant Molecular Biology, 2003,51(2):165-174.
doi: 10.1023/a:1021142403856 pmid: 12602875 |
| [19] |
刘丽, 王静, 张志明, 等. 玉米丝氨酸羧肽酶基因(ZmSCP)的克隆及表达分析. 作物学报, 2013,39(1):164-171.
doi: 10.3724/SP.J.1006.2013.00164 |
| [20] |
Liu H Z, Wang X E, Zhang H J, et al. A rice serine carboxypeptidase-like gene OsBISCPL1 is involved in regulation of defense responses against biotic and oxidative stress. Gene, 2008,420:57-65.
pmid: 18571878 |
| [1] | 李颜方,杜艳伟,张正,王高鸿,赵根有,赵晋锋,余爱丽. 农杆菌介导谷子成熟胚遗传转化体系的建立与优化[J]. 作物杂志, 2019, (3): 73–79 |
| [2] | 罗海斌, 蒋胜理, 黄诚梅, 曹辉庆, 邓智年, 吴凯朝, 徐林, 陆珍, 魏源文. 甘蔗ScHAK10基因克隆及表达分析[J]. 作物杂志, 2018, (4): 53–61 |
| [3] | 张微,王良群,刘勇,郝艳芳,杨伟,白鸿雁,武擘. 农杆菌介导高粱遗传转化的相关因素优化[J]. 作物杂志, 2018, (1): 56–61 |
| [4] | 杨浩,朱延明. 耐碱GsCHX19基因的克隆及对苜蓿的遗传转化[J]. 作物杂志, 2016, (3): 37–44 |
| [5] | 孙鑫, 杨在东, 虞光辉, 等. Pinam、Pinbm和Gsp-1m基因表达载体构建及遗传转化[J]. 作物杂志, 2015, (4): 55–60 |
| [6] | 温昕, 邵铁梅, 柴锡庆, 等. 基于组织培养的红花生物反应器研究进展[J]. 作物杂志, 2015, (1): 25–30 |
| [7] | 李兴欣, 孟义江, 罗婉娇, 等. 基于绿色荧光标记的甘草遗传转化体系的建立[J]. 作物杂志, 2014, (4): 52–58 |
| [8] | 王慧, 杨小艳, 翁建峰, 等. 抗玉米螟基因GrylA.301原核表达和玉米遗传转化[J]. 作物杂志, 2014, (2): 34–38 |
| [9] | 刘建伟, 陈晓峰, 刘广富, 郭宗端, 李新柱, 胡兆平, 张亮. 大豆CYP78A5基因组织特异性启动子的克隆及表达分析[J]. 作物杂志, 2014, (1): 54–58 |
| [10] | 向鹏, 龙承波, 罗红丽. 水稻OsMED7基因的克隆及表达分析[J]. 作物杂志, 2013, (3): 21–24 |
| [11] | 孙方志, 王大庆, 刘慧民. 小黑杨APETALA1基因的克隆与花芽发育中的表达模式[J]. 作物杂志, 2012, (4): 34–40 |
| [12] | 王伟, 王罡, 季静, 郑丽红, 关春峰. 大豆子叶节植株再生体系的优化及转EPSPS基因的研究[J]. 作物杂志, 2012, (3): 23–27 |
| [13] | 张玉, 蒋欣梅, 于锡宏. 农杆菌介导的ICE1基因转化番茄的研究[J]. 作物杂志, 2010, (5): 51–55 |
| [14] | 史冬燕, 黄兴奇. 东乡野生稻STK抗病基因片段的克隆及序列分析[J]. 作物杂志, 2009, (2): 26–29 |
| [15] | 卢翠华, 邸宏, 石瑛, 等. 马铃薯微型薯外植体遗传转化体系的优化[J]. 作物杂志, 2009, (1): 31–35 |
|