作物杂志,2025, 第5期: 102–112 doi: 10.16035/j.issn.1001-7283.2025.05.014
玉米亚油酸合成相关基因的挖掘及转录调控机制研究
杨珂( ), 赖上坤, 金倩, 王卫军, 丁翊东, 刘晓飞(
), 赖上坤, 金倩, 王卫军, 丁翊东, 刘晓飞( )
)
江苏省农业科学院宿迁农科所, 223800, 江苏宿迁
Study on the Mining and Transcriptional Regulation Mechanism of Genes Related to Linoleic Acid Synthesis in Maize
Yang Ke( ), Lai Shangkun, Jin Qian, Wang Weijun, Ding Yidong, Liu Xiaofei(
), Lai Shangkun, Jin Qian, Wang Weijun, Ding Yidong, Liu Xiaofei( )
)
Suqian Institute of Agricultural Sciences ,Jiangsu Academy of Agricultural Sciences Suqian 223800, Jiangsu, China
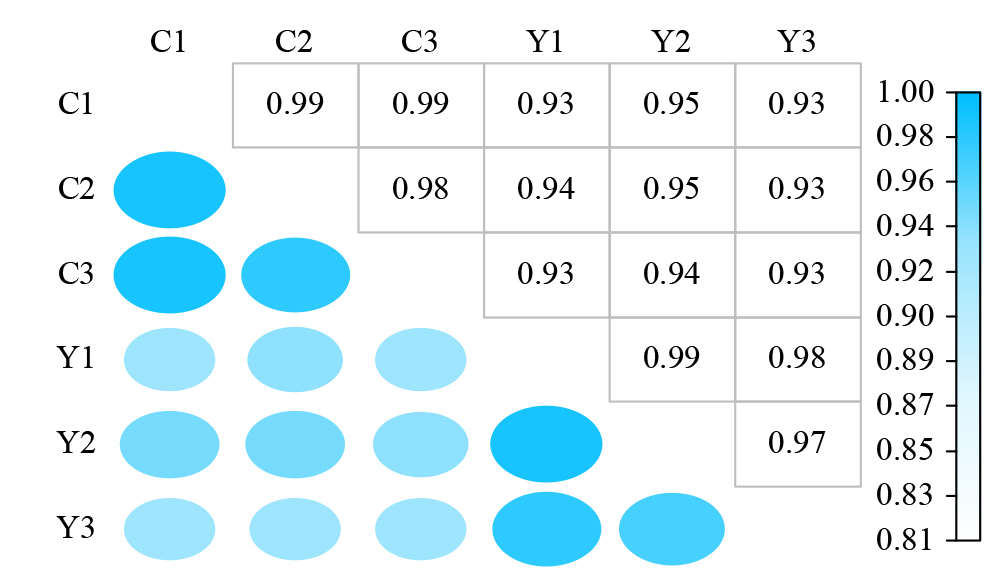
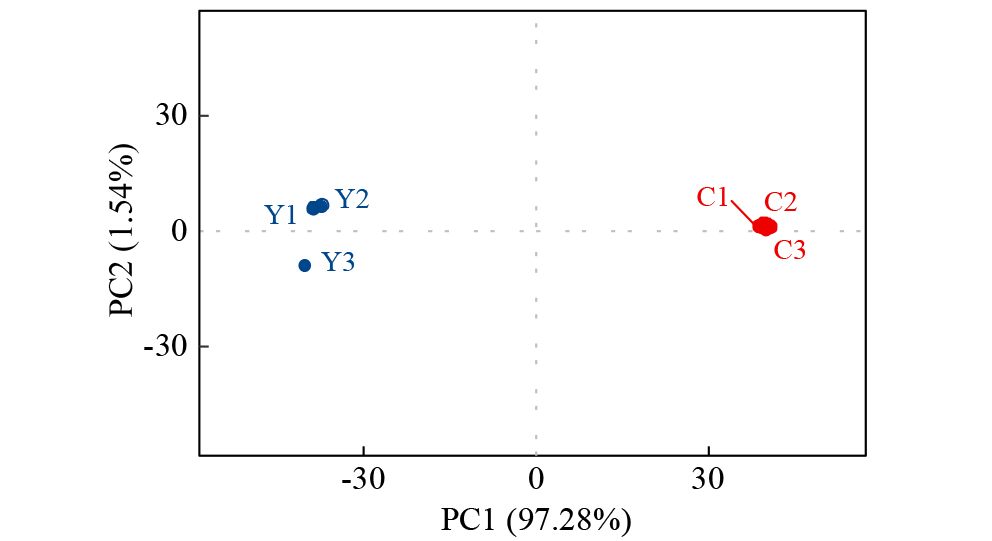
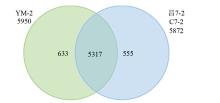
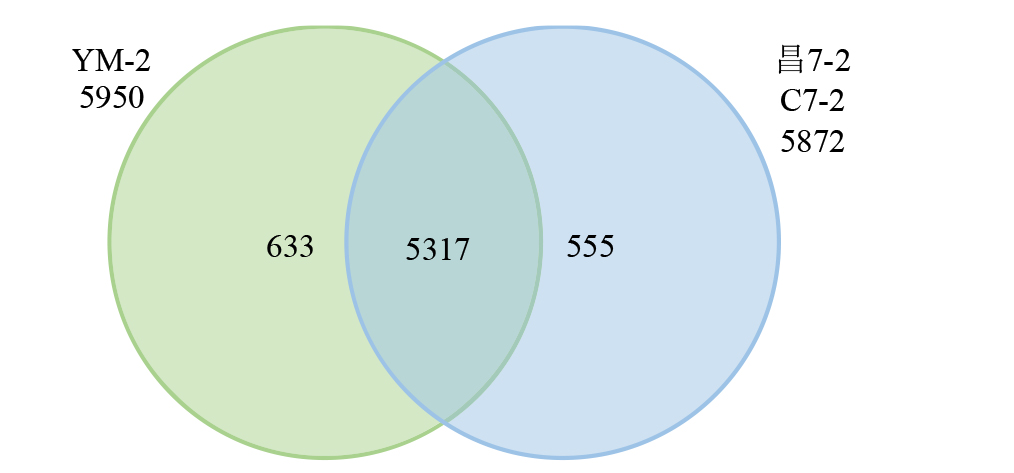
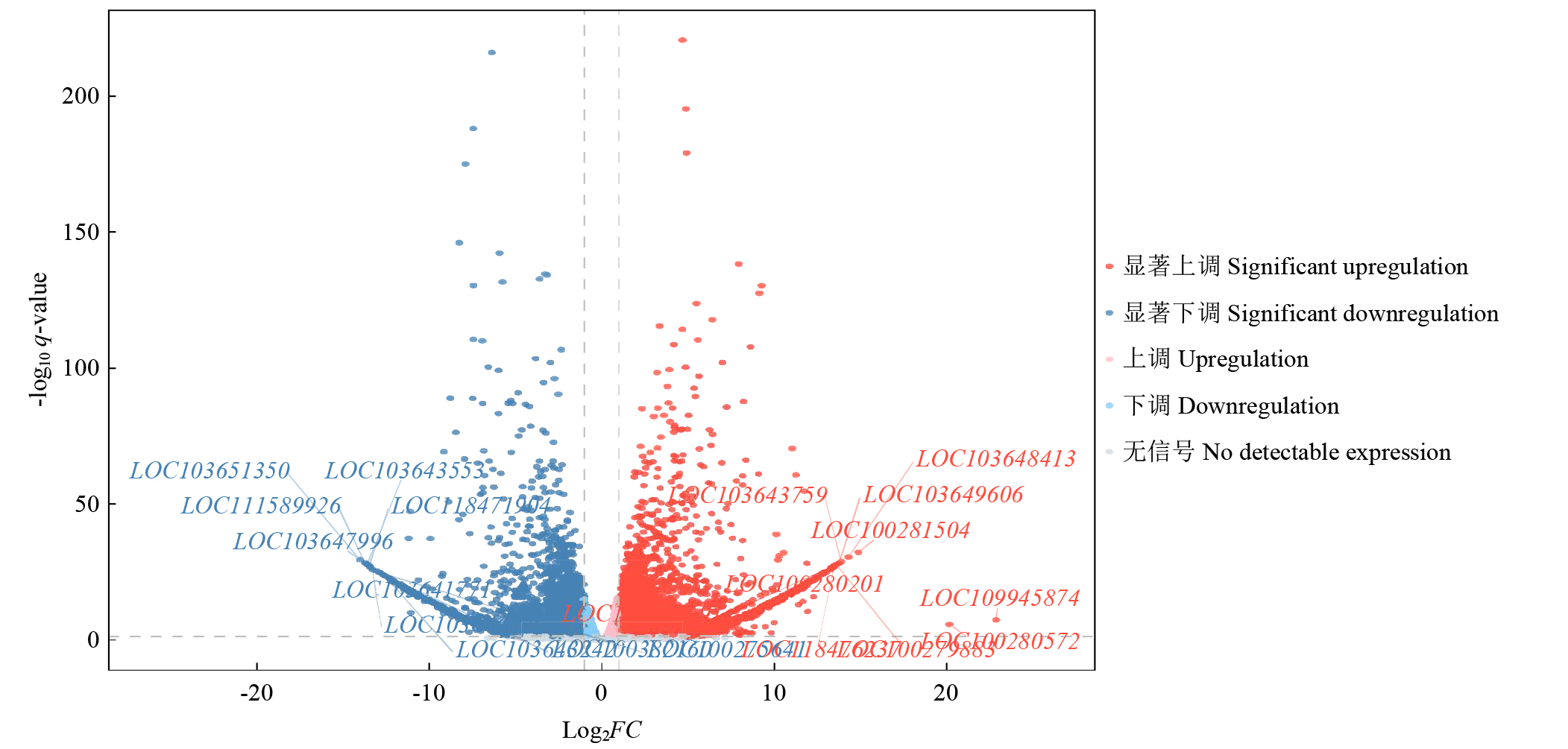
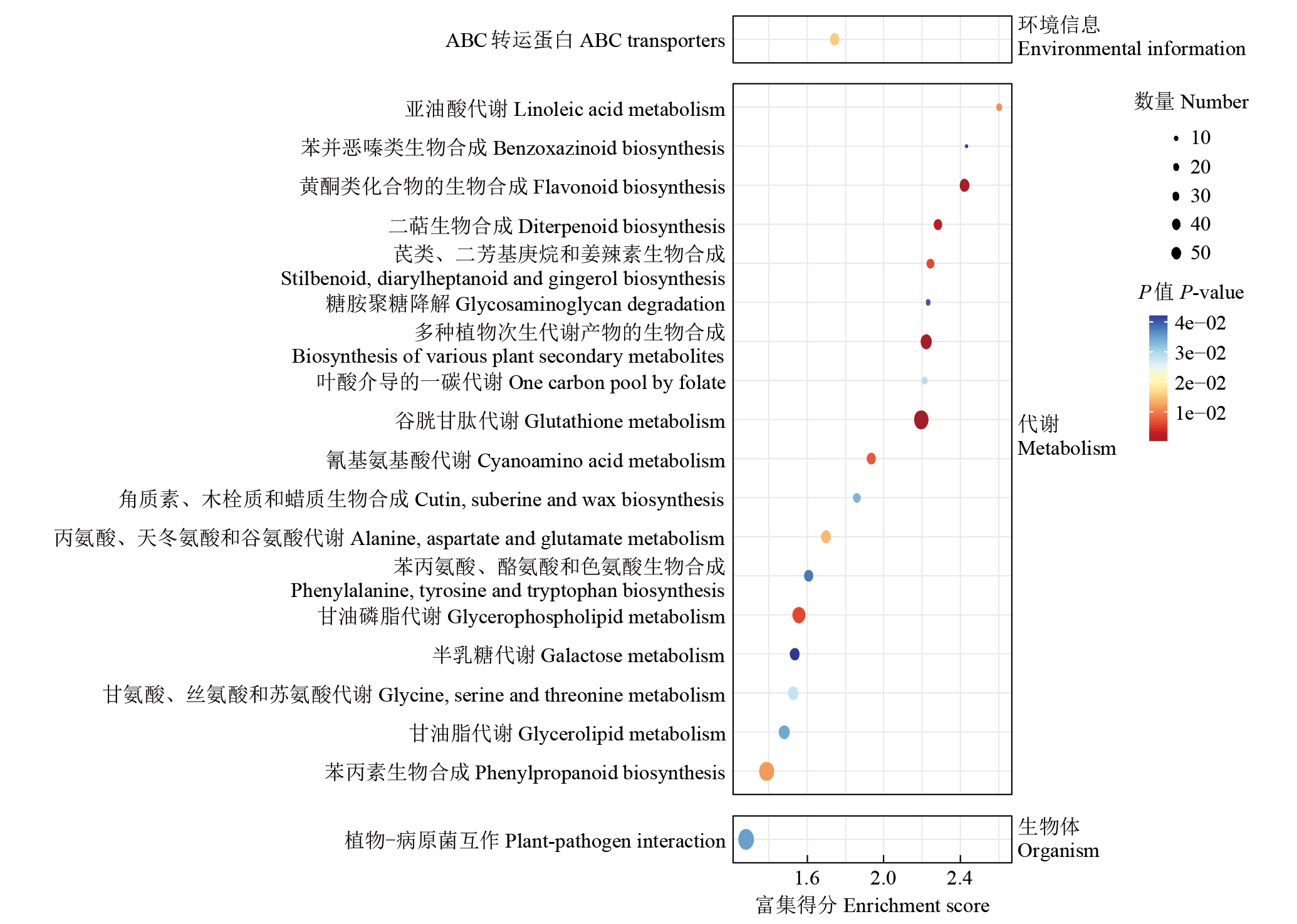
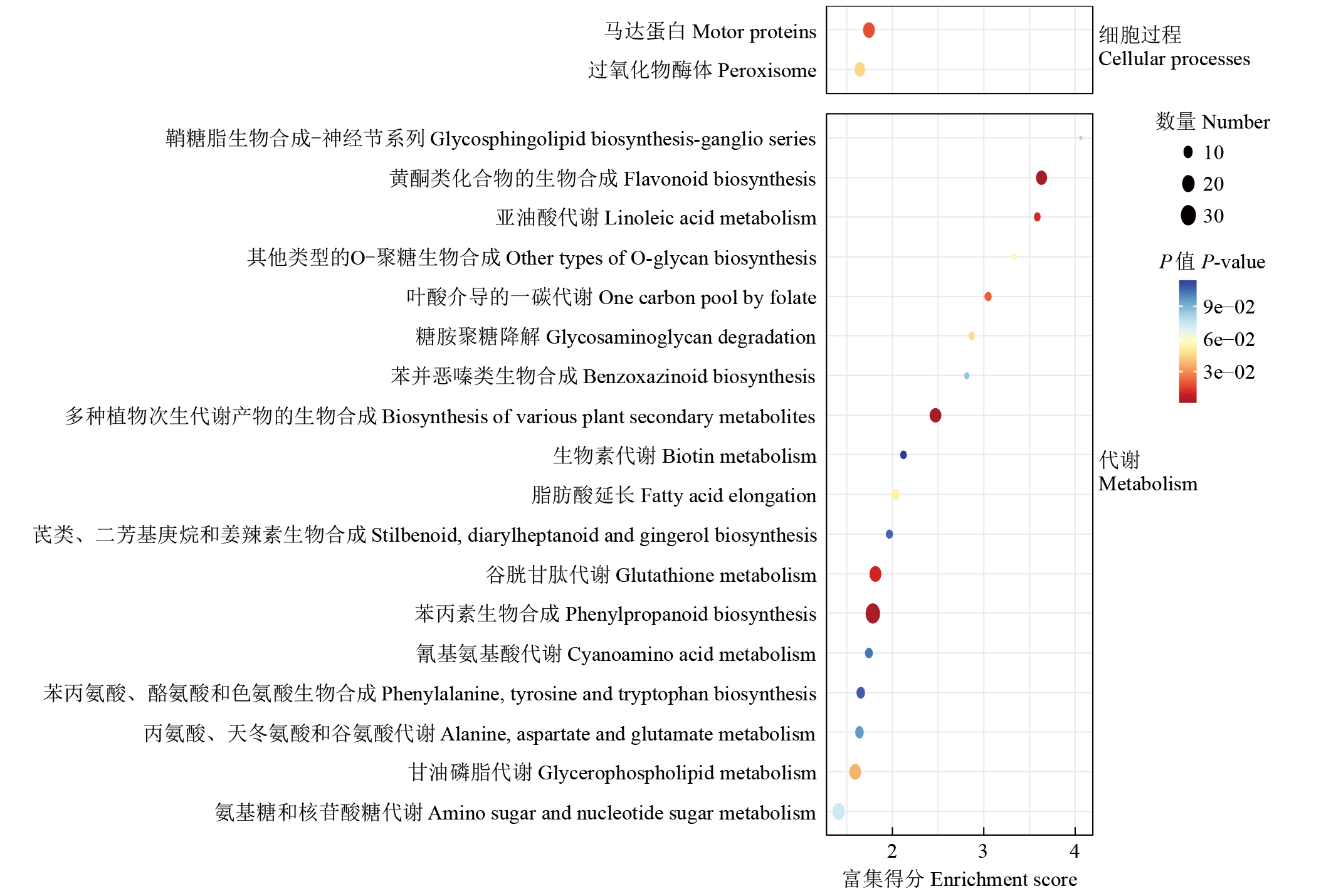
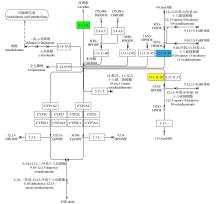
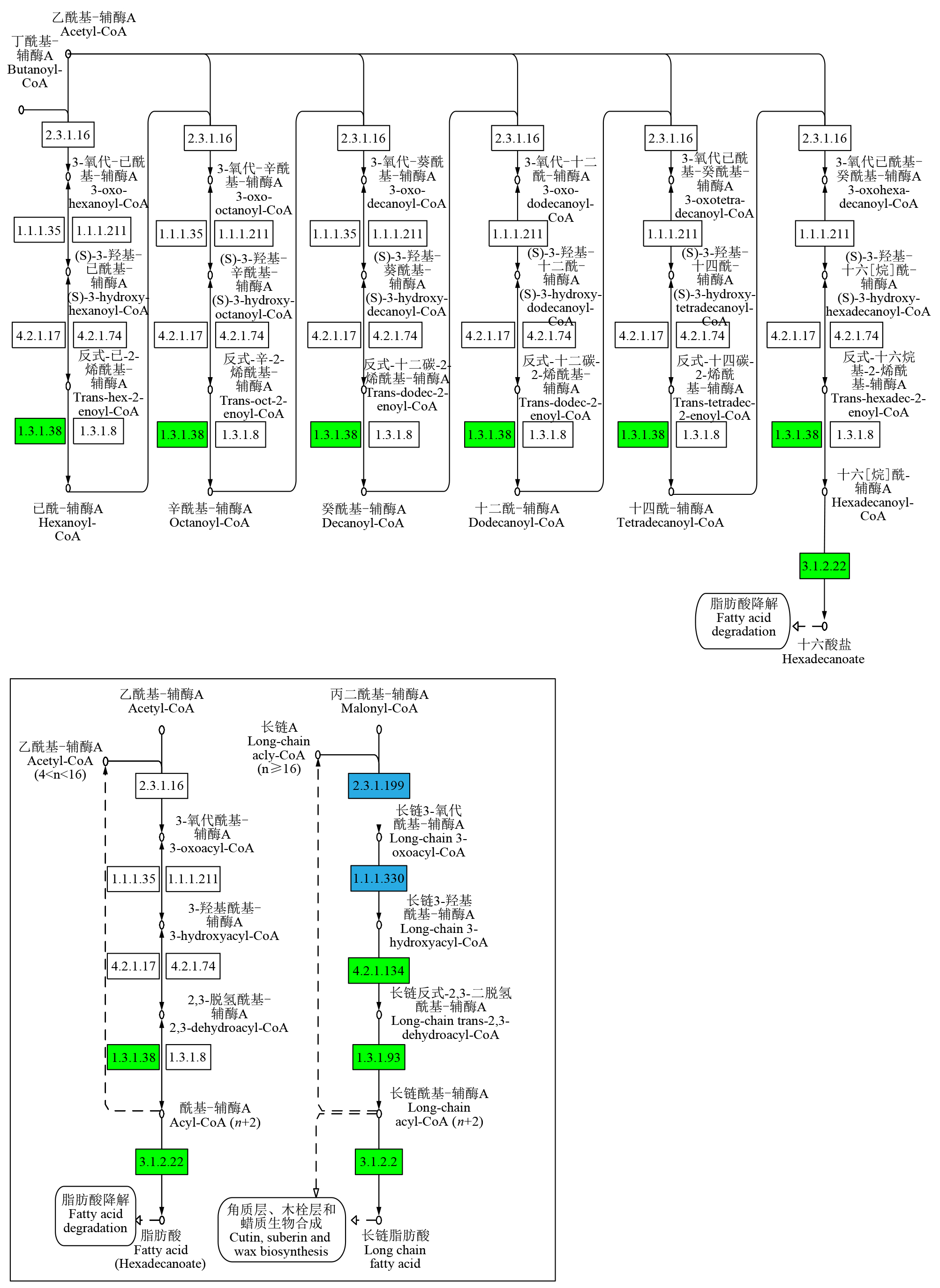
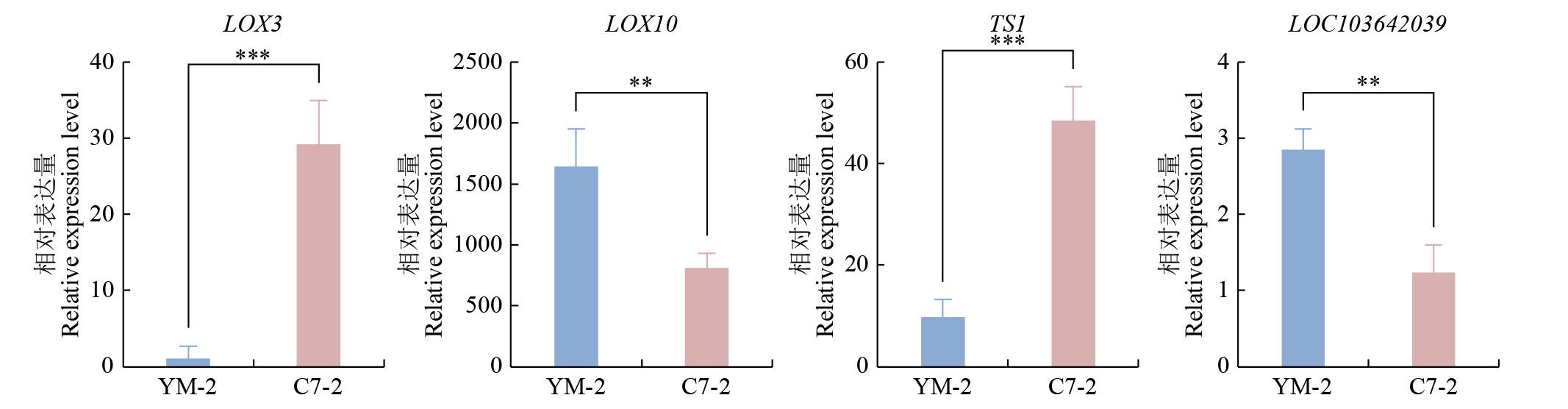
摘要: 亚油酸作为不饱和脂肪酸,对人体健康至关重要。以自主育成的高亚油酸玉米自交系YM-2和普通玉米自交系昌7-2为试验材料进行转录组学分析,挖掘玉米亚油酸生物合成相关候选基因和关键通路。结果表明,2个比较组共鉴定到6505个差异表达基因(differentially expressed genes,DEGs),其中3381个基因上调表达,3124个基因下调表达。KEGG富集分析表明,DEGs被显著富集在亚油酸代谢(Zma00591)和脂肪酸延长(Zma00062)等代谢途径中。其中,亚油酸9S-脂氧合酶(EC:1.13.11.58)、亚油酸13S-脂氧合酶(EC:1.13.11.12)、3-酮脂酰-CoA合酶(EC:2.3.1.199)和17β-雌二醇17-脱氢酶/极长链3-氧代酰基辅酶A还原酶(EC:1.1.1.62/ 1.1.1.330)表达下调,相关候选基因LOX1、LOX3、LOX6、LOX10、LOX12、TS1、LOC103642039和LOC103646607可能与亚油酸的合成直接相关。为验证RNA-seq数据的准确性,选取在2份材料中表达模式相反的4个基因进行qRT-PCR验证,发现基因表达量的变化趋势与转录组测序结果一致。本研究挖掘玉米亚油酸生物合成的相关基因和代谢通路,为深入解析玉米高亚油酸的分子调控机制提供参考,也为高油酸农作物的分子育种奠定基础。
| [1] | 倪雪梅, 胡玉兰, 李旭, 等. 大胚高油玉米籽粒蛋白质含量相关QTLs定位研究. 分子植物育种,(2024-03-07)[2025-04-18]. https://link.cnki.net/urlid/46.1068.S.20240305.1751.018. |
| [2] | 刘晓飞, 崔小平, 孙宝林, 等. 高亚油酸玉米新组合生长发育特性及亚油酸含量. 江西农业学报, 2019, 31(11):11-16. |
| [3] |
Lacombe R J S. Chouinard-Watkins R, Bazinet R P. Brain docosahexaenoic acid uptake and metabolism. Molecular Aspects of Medicine, 2018, 64:109-134.
doi: S0098-2997(17)30135-8 pmid: 29305120 |
| [4] | 戴永军. 日粮亚油酸对中华绒螯蟹生长、脂氧合酶代谢和风味品质的影响. 南京:南京农业大学, 2022. |
| [5] | 姚池蓓. 不同比例的α-亚麻酸/亚油酸对生长后期草鱼生产性能和肉质的影响及其作用机制. 雅安:四川农业大学, 2023. |
| [6] |
Li H, Peng Z Y, Yang X H, et al. Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nature Genetics, 2013, 45(1):43-50.
doi: 10.1038/ng.2484 pmid: 23242369 |
| [7] | 杨凤萍, 李向龙, 张立全, 等. 双功能去饱和酶AcD12基因改良玉米不饱和脂肪酸成分. 分子植物育种, 2017, 15(12):4805-4812. |
| [8] |
Shen B, Allen W B, Zheng P Z, et al. Expression of ZmLEC1 and ZmWRI1 increases seed oil production in maize. Plant Physiology, 2010, 153(3):980-987.
doi: 10.1104/pp.110.157537 pmid: 20488892 |
| [9] | Wang J B, Liu L, Zhang Q, et al. Genome-wide association analysis-based mining of quality genes related to linoleic and linolenic acids in soybean. Agriculture, 2023, 13:2250. |
| [10] | Di Q, Piersanti A, Zhang Q, et al. Genome-wide association study identifies candidate genes related to the linoleic acid content in soybean seeds. International Journal of Molecular Sciences, 2021, 23(1):454. |
| [11] | Wang F L, Zheng T, et al. Overexpression of miR319a altered oil body morphogenesis and lipid content in Arabidopsis seeds. Plant Molecular Biology Reporter, 2020, 38:531-537. |
| [12] | Wang T, Wang F H, Deng S H, et al. Single-cell transcriptomes reveal spatiotemporal heat stress response in maize roots. Nature Communications, 2025, 16:177. |
| [13] |
Lin Y N, et al. Utilizing transcriptomics and metabolomics to unravel key genes and metabolites of maize seedlings in response to drought stress. BMC Plant Biology, 2024, 24:34.
doi: 10.1186/s12870-023-04712-y pmid: 38185653 |
| [14] |
Song G S, et al. Gene expression and expression quantitative trait loci analyses uncover natural variations underlying the improvement of important agronomic traits during modern maize breeding. The Plant Journal, 2023, 115(3):772-787.
doi: 10.1111/tpj.16260 pmid: 37186341 |
| [15] | 郭新月, 张岚, 张晴, 等. 转录组测序解析油菜素甾醇调控番茄根发育的机制. 河北师范大学学报, 2022, 46(5):512-519. |
| [16] | 王继玥, 余庭跃, 张采波.玉米转录组学研究进展. 华北农学报 , 2014,29(增):10-15. |
| [17] | Luo M J, Shi Y X, et al. Genetic basis of the oil biosynthesis in ultra-high-oil maize grains with an oil content exceeding 20%. Front Plant Science, 2023, 14:1168216. |
| [18] | Pouvreau B, Baud S, Vernoud V, et al. Duplicate maize Wrinkled1 transcription factors activate target genes involved in seed oil biosynthesis. Plant Physiology, 2011, 156(2):674-686. |
| [19] | Kanehisa M, Araki M, Goto S, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Research,2008,36:D480-D484. |
| [20] | Ogunola O F, Hawkins L K, Mylroie E, et al. Characterization of the maize lipoxygenase gene family in relation to aflatoxin accumulation resistance. PLoS ONE, 2017, 12(7):e0181265. |
| [21] |
Acosta I F, Laparra H, Romero S P, et al. tasselseed1 is a lipoxygenase affecting jasmonic acid signaling in sex determination of maize. Science, 2009, 323(5911):262-265.
doi: 10.1126/science.1164645 pmid: 19131630 |
| [22] | Xu Y, Yan F, Liu Y J, et al. Quantitative proteomic and lipidomics analyses of high oil contentGmDGAT1-2 transgenic soybean illustrate the regulatory mechanism of lipoxygenase and oleosin. Plant Cell Report, 2021, 40(12):2303-2323. |
| [23] |
Pan R H, Liu J, Wang S S, et al. Peroxisomes: versatile organelles with diverse roles in plants. New Phytologist, 2020, 225(4):1410-1427.
doi: 10.1111/nph.16134 pmid: 31442305 |
| [24] |
Braun D M. Phloem loading and unloading of sucrose: what a long, strange trip from source to sink. Annual Review of Plant Biology, 2022, 73:553-584.
doi: 10.1146/annurev-arplant-070721-083240 pmid: 35171647 |
| [25] |
Feussner I, Wasternack C. The lipoxygenase pathway. Annual Review of Plant Biology, 2002, 53:275-297.
pmid: 12221977 |
| [26] | Gao X Q, Starr J, Göbel C, et al. Maize 9-lipoxygenase ZmLOX3 controls development, root-specific expression of defense genes, and resistance to root-knot nematodes. Molecular Plant-Microbe Interactions, 2008, 21(1):98-109. |
| [27] | Gao X Q, Brodhagen M, Isakeit T, et al. Inactivation of the lipoxygenase ZmLOX3 increases susceptibility of maize to Aspergillus spp.. Molecular Plant-Microbe Interactions, 2009, 22(2):222-231. |
| [1] | 周婷芳, 李冉, 刘倩倩, 张泽, 王振华, 马宝新, 路明, 张林, 韩业辉, 杨波, 李明顺, 张德贵, 翁建峰, 雍洪军, 徐晶宇, 韩洁楠, 李新海. 东北区118份玉米杂交种萌发期耐盐性分析[J]. 作物杂志, 2025, (5): 1–10 |
| [2] | 王昌亮, 闫丽慧, 常建智, 张国合, 王静, 王芬霞, 黄利华. 玉米新品种浚单168的选育与创新思路[J]. 作物杂志, 2025, (5): 142–146 |
| [3] | 朱建国, 崔正果, 张波, 王学端, 张月明, 吕小飞, 王洪预, 李秋祝, 崔金虎. 玉米―花生―谷子轮作对吉林省风沙盐碱地区土壤微生态、产量和效益的影响[J]. 作物杂志, 2025, (5): 19–28 |
| [4] | 刘弟, 宋来贵, 田临卿, 李志刚, 马俊美, 李作森, 年夫照, 焦健, 邓小鹏. 烟后套作玉米对作物生产和土壤养分的影响[J]. 作物杂志, 2025, (5): 239–246 |
| [5] | 李喆豪, 姬米源, 吕梦, 明博, 李少昆, 张海艳, 谢瑞芝. 长期定位条件下不同氮肥运筹对春玉米根冠发育的影响[J]. 作物杂志, 2025, (4): 135–141 |
| [6] | 王兴亚, 陈宇涵, 张孟雯, 孙琳琳, 陈利容, 郭玉秋, 龚魁杰. 不同时期施用脱落酸对玉米籽粒灌浆和脱水的影响[J]. 作物杂志, 2025, (4): 173–180 |
| [7] | 贺新春, 杜何为, 黄敏. 玉米ZmCaM1基因的克隆和表达分析[J]. 作物杂志, 2025, (4): 19–28 |
| [8] | 吴凤婕, 侯楠, 齐翔鲲, 杨克军, 付健, 王玉凤. 不同施氮量对半干旱区糯玉米主要营养品质及产量的影响[J]. 作物杂志, 2025, (4): 267–275 |
| [9] | 史亚兴, 刘俊玲, 朱贵川, 赫忠友, 刘辉, 樊艳丽, 徐丽, 卢柏山, 赵久然, 骆美洁. du1新等位变异的克隆及其分子标记的开发[J]. 作物杂志, 2025, (4): 41–48 |
| [10] | 陈明辉, 徐豫, 黄志强, 王俊青, 张保青. 冷胁迫下果蔗叶片转录组分析及茉莉酸信号传导基因挖掘[J]. 作物杂志, 2025, (4): 95–103 |
| [11] | 侯岳, 王红亮, 李杰, 李春杰, 陈范骏. 禾本科与豆科饲草作物间套作对饲草品质及氮素吸收影响的研究进展[J]. 作物杂志, 2025, (3): 1–10 |
| [12] | 金梓浩, 赵文清, 王芳, 王威, 彭云玲, 常芳国. 三种外源植物生长调节物质对玉米幼苗耐寒性的影响[J]. 作物杂志, 2025, (3): 125–132 |
| [13] | 王祎, 任永福, 张正鹏, 丁德芳, 张靖, 刘祎鸿, 孙多鑫, 陈光荣. 不同覆盖材料对河西灌区土壤环境及玉米产量的影响[J]. 作物杂志, 2025, (3): 149–155 |
| [14] | 侯楠, 吴凤婕, 齐翔鲲, 王玉凤, 杨克军, 付健. 不同施氮水平对黑土区糯玉米灌浆期碳代谢的影响[J]. 作物杂志, 2025, (3): 178–184 |
| [15] | 李洪涛, 柴文波, 许瀚元, 李淑芬, 祝庆, 袁超, 王军. 不同鲜食糯玉米品种货架期评价及其鉴定指标筛选[J]. 作物杂志, 2025, (3): 241–248 |
|
||