作物杂志,2025, 第5期: 135–141 doi: 10.16035/j.issn.1001-7283.2025.05.018
水稻CYP450家族基因Os78A5的表达分析
植藓闳1( ), 纪子贤1, 许镇旺1, 谭恩1, 梁日深1, 马帅鹏2, 唐辉武1(
), 纪子贤1, 许镇旺1, 谭恩1, 梁日深1, 马帅鹏2, 唐辉武1( )
)
- 1
仲恺农业工程学院农业与生物学院, 510225, 广东广州
2中国热带农业科学院广州实验站, 510145, 广东广州
Expression Analysis of the CYP450 Family Gene Os78A5 in Rice
Zhi Xianhong1( ), Ji Zixian1, Xu Zhenwang1, Tan En1, Liang Rishen1, Ma Shuaipeng2, Tang Huiwu1(
), Ji Zixian1, Xu Zhenwang1, Tan En1, Liang Rishen1, Ma Shuaipeng2, Tang Huiwu1( )
)
- 1
College of Agriculture and Biology ,Zhongkai University of Agriculture and Engineering Guangzhou 510225, Guangdong, China
2Guangzhou Experimental Station ,Chinese Academy of Tropical Agricultural Sciences Guangzhou 510145, Guangdong, China
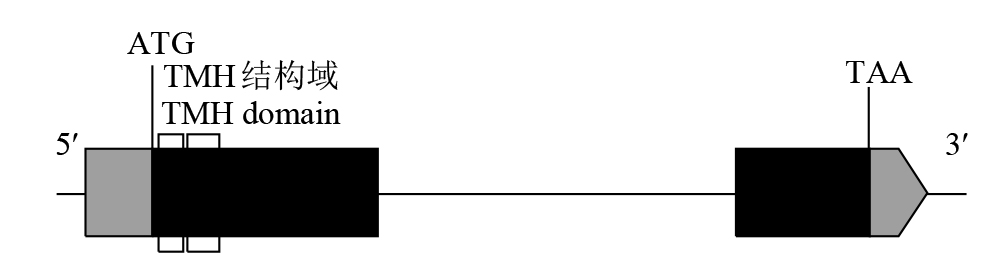
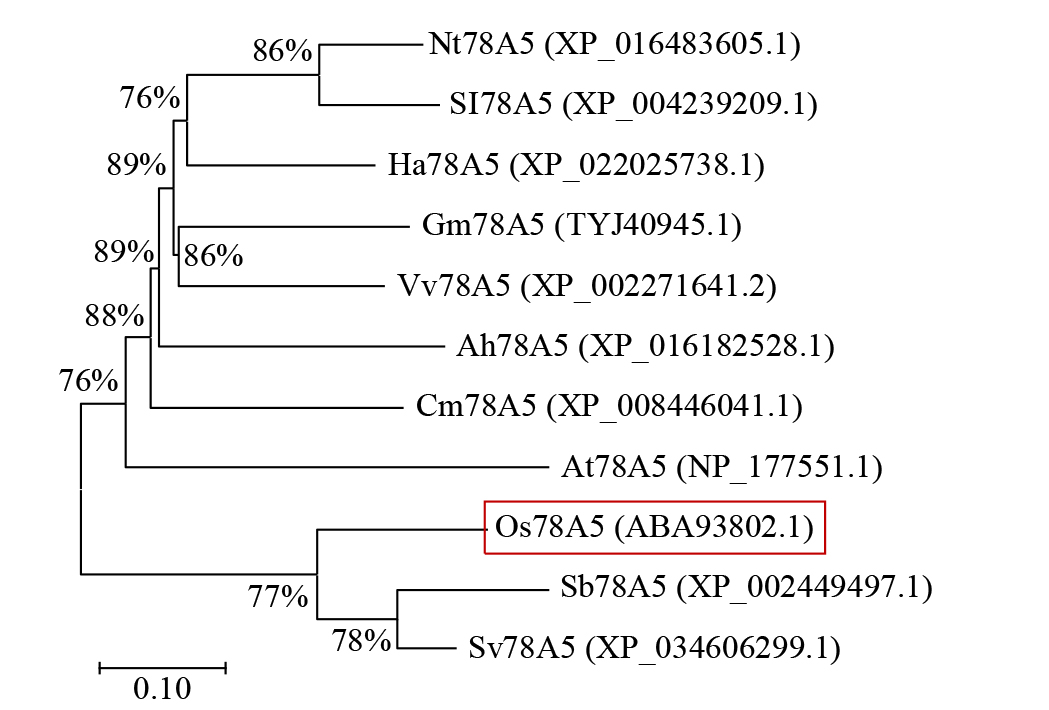
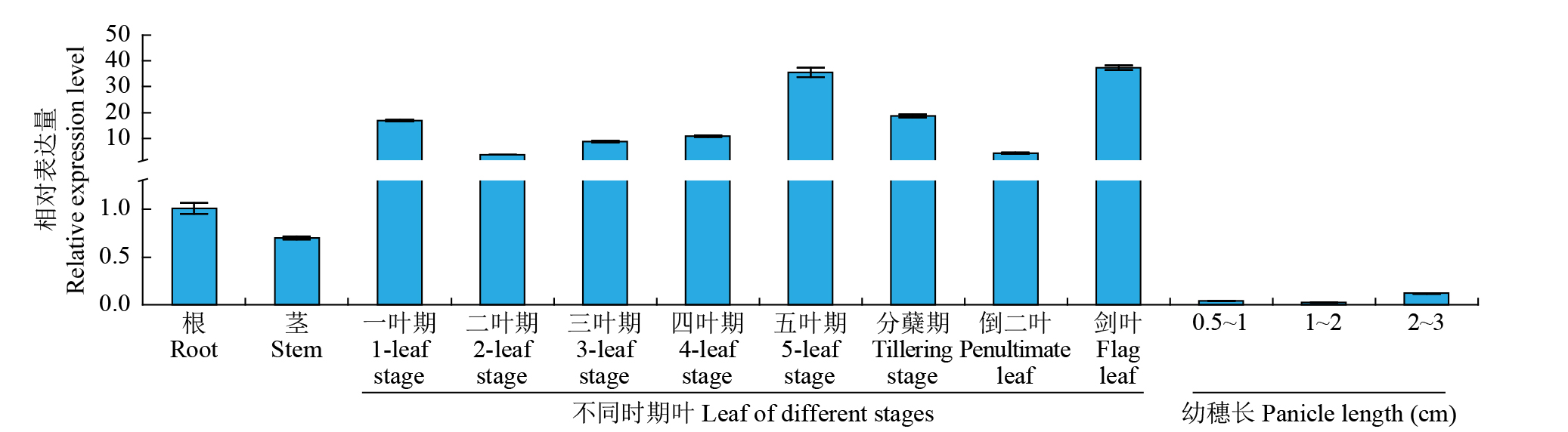
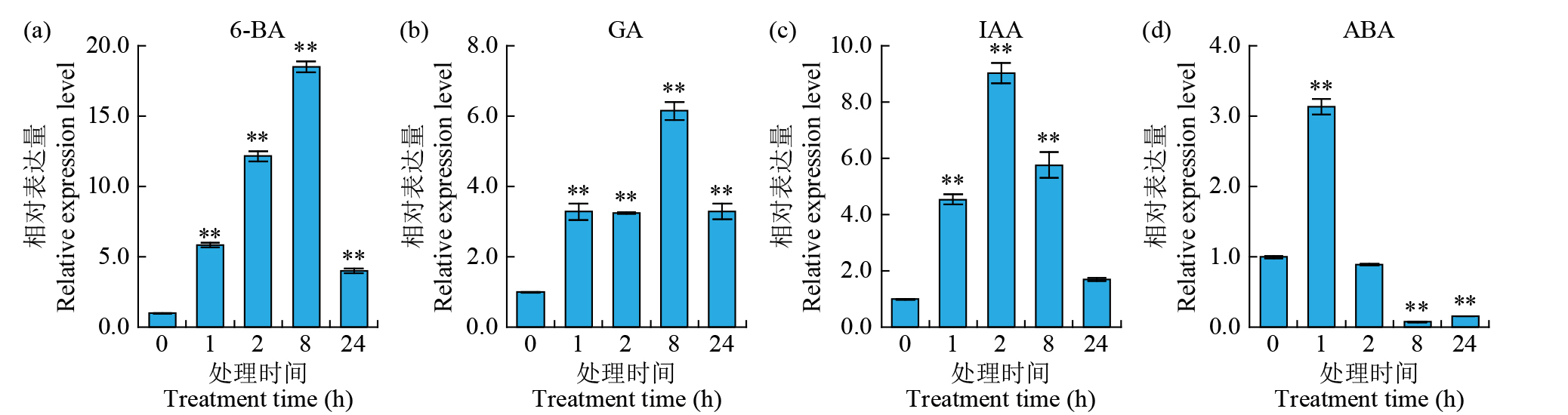
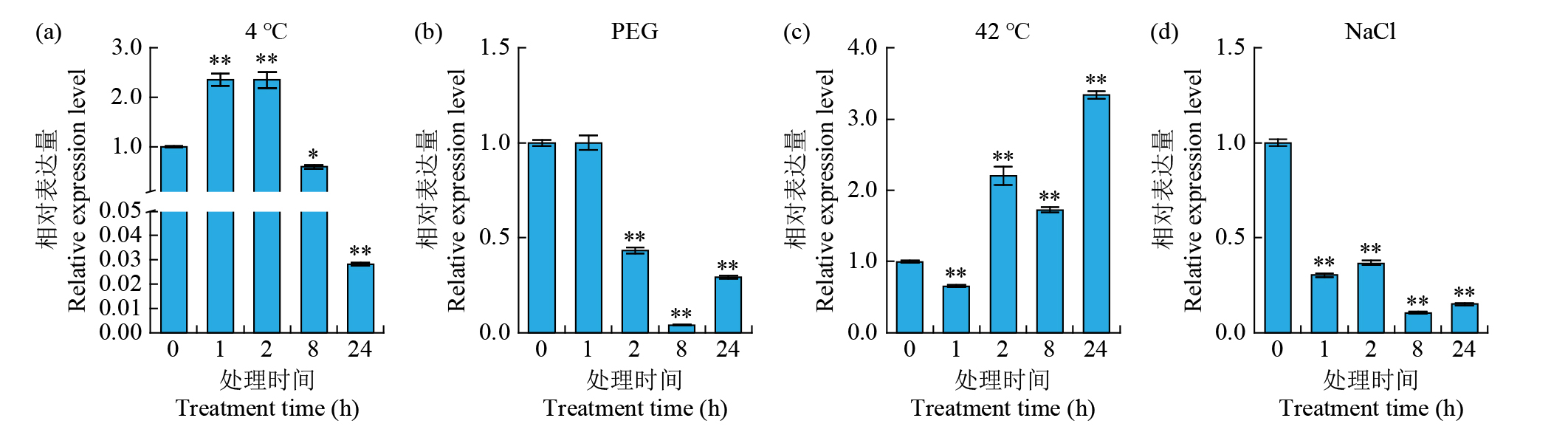
摘要: 细胞色素P450(cytochrome P450,CYP450)广泛存在于各种生物中,在植物次生代谢和胁迫响应过程中具有不可或缺的作用。在水稻(Oryza sativa)中克隆了一个编码CYP450蛋白的基因Os78A5(LOC_Os11g29720),并利用生物信息学技术分析Os78A5基因结构、演化过程和顺式作用元件,同时利用实时荧光定量PCR(quantitative real-time PCR,qRT-PCR)技术分析Os78A5的组织表达特异性及其对不同激素和非生物胁迫的响应情况。结果表明,Os78A5编码区长度1617 bp,编码538个氨基酸。Os78A5与狗尾草(Setaria viridis)和高粱(Sorghum bicolor)的同源蛋白CYP78A5具有较近的亲缘关系。Os78A5的启动子区包含48个激素响应元件、23个环境胁迫响应元件和35个水分响应顺式作用元件。Os78A5在水稻叶片中的表达水平显著高于其他组织,且受细胞分裂素、赤霉素、生长素、脱落酸、高温和低温等诱导表达,推测Os78A5可能参与水稻的非生物胁迫响应。
| [1] | 杨杰, 詹亚光, 肖佳雷, 等. 细胞色素 P450 在植物三萜和甾醇骨架修饰中的功能研究进展. 中国科学:生命科学, 2018, 48(10):1065-1083. |
| [2] |
Guengerich F P. Cytochrome P450 research and the journal of biological chemistry. Journal of Biological Chemistry, 2019, 294(5):1671-1680.
doi: 10.1074/jbc.TM118.004144 pmid: 29871932 |
| [3] | Nelson D R. Cytochrome P450 diversity in the tree of life. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics, 2018, 1866(1):141-154. |
| [4] |
Nelson D, Werck‐Reichhart D. A P450‐centric view of plant evolution. The Plant Journal, 2011, 66(1):194-211.
doi: 10.1111/j.1365-313X.2011.04529.x pmid: 21443632 |
| [5] | Bak S, Beisson F, Bishop G, et al. Cytochromes P450. The Arabidopsis Book, 2011, 9:e0144. |
| [6] | 罗浑金, 傅童声. 细胞色素P450的多样性与介导抗性的研究. 生物技术通报, 2006(2):14-16. |
| [7] | Chapple C. Molecular-genetic analysis of plant cytochrome P450- dependent monooxygenases. Annual Review of Plant Biology, 1998, 49(1):311-343. |
| [8] | Wang Y F, Ahmad N, et al. Unraveling the functional characterization of a jasmonate-induced flavonoid biosynthetic CYP45082G24 gene in Carthamus tinctorius. Functional & Integrative Genomics, 2023, 23(2):172. |
| [9] | Wang H Y, Wang Y, et al. Genome-wide identification of the CYP82 gene family in cucumber and functional characterization of CsCYP82D102 in regulating resistance to powdery mildew.. PeerJ, 2024, 12:e17162. |
| [10] |
He J, Chen Q W, Xin P Y, et al. CYP72A enzymes catalyse 13-hydrolyzation of gibberellins. Nature Plants, 2019, 5(10):1057-1065.
doi: 10.1038/s41477-019-0511-z pmid: 31527846 |
| [11] | Pandian B A, Sathishraj R, Djanaguiraman M, et al. Role of cytochrome P450 enzymes in plant stress response. Antioxidants, 2020, 9(5):454. |
| [12] | Rao M J, Xu Y, Tang X, et al. CsCYT75B1, a citrus CYTOCHROME P 450 gene, is involved in accumulation of antioxidant flavonoids and induces drought tolerance in transgenic Arabidopsis. Antioxidants, 2020, 9(2):161. |
| [13] | Yan Q, Cui X X, Lin S, et al. GmCYP82A3,a soybean cytochrome P 450 family gene involved in the jasmonic acid and ethylene signaling pathway, enhances plant resistance to biotic and abiotic stresses. PLoS ONE, 2016, 11(9):e0162253. |
| [14] |
Jiang L, Yoshida T, Stiegert S, et al. Multi-omics approach reveals the contribution of KLU to leaf longevity and drought tolerance. Plant Physiology, 2021, 185(2):352-368.
doi: 10.1093/plphys/kiaa034 pmid: 33721894 |
| [15] | 李泽宇.OsCYP71D8L 调控水稻株高和耐逆性的分子基础. 北京: 中国农业科学院, 2021. |
| [16] | Kurotani K, Hayashi K, Hatanaka S, et al. Elevated levels of CYP94 family gene expression alleviate the jasmonate response and enhance salt tolerance in rice. Plant and Cell Physiology, 2015, 56(4):779-789. |
| [17] | Maeda S, Dubouzet J G, Kondou Y, et al. The rice CYP78A gene BSR2 confers resistance to Rhizoctonia solani and affects seed size and growth in Arabidopsis and rice. Scientific Reports, 2019, 9(1):587. |
| [18] | Wang A J, Ma L, Shu X Y, et al. Rice (Oryza sativa L.) cytochrome P 450 protein 716A subfamily CYP716A16 regulates disease resistance. BMC Genomics, 2022, 23(1):343. |
| [19] |
Sakamoto T, Kawabe A, Tokida‐Segawa A, et al. Rice CYP734As function as multisubstrate and multifunctional enzymes in brassinosteroid catabolism. The Plant Journal, 2011, 67(1):1-12.
doi: 10.1111/j.1365-313X.2011.04567.x pmid: 21418356 |
| [20] | Zhou C L, Lin Q B, Ren Y L, et al. A CYP78As-small grain4-coat protein complex Ⅱ pathway promotes grain size in rice. The Plant Cell, 2023, 35(12):4325. |
| [21] | Sahoo B, Nayak I, Parameswaran C, et al. A comprehensive genome-wide investigation of the cytochrome 71 (OsCYP71) gene family: revealing the impact of promoter and gene variants (Ser33Leu) of OsCYP71P6 on yield-related traits in indica rice (Oryza sativa L.). Plants, 2023, 12(17):3035. |
| [22] | Song S F, Wang T K, et al. A novel strategy for creating a new system of third‐generation hybrid rice technology using a cytoplasmic sterility gene and a genic male‐sterile gene. Plant Biotechnology Journal, 2021, 19(2):251-260. |
| [23] | Li H, Pinot F, Sauveplane V, et al. Cytochrome P450 family member CYP704B2 catalyzes the ω-hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. The Plant Cell, 2010, 22(1):173-190. |
| [24] | Huang X Y, Chang Z Y, et al. Regulation by distinct MYB transcription factors defines the roles of OsCYP86A9 in anther development and root suberin deposition. The Plant Journal, 2024, 118(6):1972-1990. |
| [25] | Cui Y J, Peng Y L, Zhang Q, et al. Disruption of EARLY LESION LEAF 1, encoding a cytochrome P 450 monooxygenase, induces ROS accumulation and cell death in rice. The Plant Journal, 2021, 105(4):942-956. |
| [26] | Berens M L, Wolinska K W, Spaepen S, et al. Balancing trade- offs between biotic and abiotic stress responses through leaf age- dependent variation in stress hormone cross-talk. Proceedings of the National Academy of Sciences of the United States of America, 2019, 116(6):2364-2373. |
| [27] |
Mafu S, Ding Y, Murphy K M, et al. Discovery, biosynthesis and stress-related accumulation of dolabradiene-derived defenses in maize. Plant Physiology, 2018, 176(4):2677-2690.
doi: 10.1104/pp.17.01351 pmid: 29475898 |
| [28] |
Xu J, Wang X Y, Guo W Z. The cytochrome P450 superfamily: Key players in plant development and defense. Journal of Integrative Agriculture, 2015, 14(9):1673-1686.
doi: 10.1016/S2095-3119(14)60980-1 |
| [29] |
Tao X, Wang M X, Dai Y, et al. Identification and expression profile of CYPome in perennial ryegrass and tall fescue in response to temperature stress. Frontiers in Plant Science, 2017, 8:1519.
doi: 10.3389/fpls.2017.01519 pmid: 29209335 |
| [30] | Wang M, Yuan J R, Qin L M, et al. TaCYP81D5, one member in a wheat cytochrome P 450 gene cluster, confers salinity tolerance via reactive oxygen species scavenging. Plant Biotechnology Journal, 2020, 18(3):791-804. |
| [31] | Kajino T, Yamaguchi M, Oshima Y, et al. KLU/CYP78A5, a cytochrome P450 monooxygenase identified via fox hunting, contributes to cuticle biosynthesis and improves various abiotic stress tolerances Frontiers in Plant Science, 2022, 13:904121. |
| [32] |
Zhang D, Yang H F, Wang X C, et al. Cytochrome P450 family member CYP96B5 hydroxylates alkanes to primary alcohols and is involved in rice leaf cuticular wax synthesis. New Phytologist, 2020, 225(5):2094-2107.
doi: 10.1111/nph.16267 pmid: 31618451 |
| [33] |
王家利, 刘冬成, 郭小丽, 等. 生长素合成途径的研究进展. 植物学报, 2012, 47(3):292-301.
doi: 10.3724/SP.J.1259.2012.00292 |
| [34] | Zhao Y, Hull A K, Gupta N R, et al. Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes & Development, 2002, 16(23):3100-3112. |
| [35] | Tamiru M, Undan J R, Takagi H, et al. A cytochrome P450, OsDSS1, is involved in growth and drought stress responses in rice (Oryza sativa L.). Plant Molecular Biology, 2015, 88:85-99. |
| [36] | Kushiro T, Okamoto M, Nakabayashi K, et al. The Arabidopsis cytochrome P 450 CYP707A encodes ABA 8′‐hydroxylases: key enzymes in ABA catabolism. The EMBO Journal, 2004, 23(7):1647-1656. |
| [37] |
Sasaki E, Ogura T, Takei K, et al. Uniconazole, a cytochrome P450 inhibitor, inhibits trans-zeatin biosynthesis in Arabidopsis. Phytochemistry, 2013, 87:30-38.
doi: 10.1016/j.phytochem.2012.11.023 pmid: 23280040 |
| [1] | 滕文, 叶凡, 周舟, 王屿乐, 刘立军. 小麦和油菜秸秆还田处理对盐胁迫下水稻产量和品质的影响[J]. 作物杂志, 2025, (5): 11–18 |
| [2] | 彭彬峰, 陆楚盛, 尹媛红, 朱菲菲, 叶群欢, 潘俊峰, 刘彦卓, 胡香玉, 胡锐, 李妹娟, 王昕钰, 梁开明, 傅友强. 高温胁迫下铵硝混配营养促进水稻生长的生理机制[J]. 作物杂志, 2025, (5): 165–170 |
| [3] | 都晗萌, 陈雨琼, 刘若彤, 陈英龙, 戴其根, 张洪程, 廖萍. 盐胁迫下喷施矮壮素和施用石膏对水稻产量及倒伏特性的影响[J]. 作物杂志, 2025, (5): 29–34 |
| [4] | 杨林生, 习敏, 涂德宝, 李忠, 周永进, 许有尊, 孙雪原, 吴文革. 长江三角洲地区主要类型水稻生产资源投入及碳、氮足迹评估[J]. 作物杂志, 2025, (4): 150–156 |
| [5] | 贺新春, 杜何为, 黄敏. 玉米ZmCaM1基因的克隆和表达分析[J]. 作物杂志, 2025, (4): 19–28 |
| [6] | 陶祖豪, 王慰亲, 郑华斌, 向军, 唐启源. 水肥及化控对晚稻有序机抛秧秧苗素质的影响[J]. 作物杂志, 2025, (4): 224–230 |
| [7] | 补清, 杨开强, 王添, 施思, 信国琛, 陈勇, 董朝霞, 安静. 纳米材料使用方法及缓解作物非生物胁迫的研究进展[J]. 作物杂志, 2025, (4): 9–18 |
| [8] | 赫兵, 王晓航, 李超, 罗立强, 张强, 韩康顺, 陈殿元, 严光彬, 刘振蛟. 1987-2022年吉林省水稻审定品种数据分析[J]. 作物杂志, 2025, (3): 16–22 |
| [9] | 曹正男, 赵振东, 胡博, 于涵, 宁晓海, 赵泽强, 曹立勇. 氮肥与促腐菌肥配施对寒地水稻秸秆还田腐解效果及产量的影响[J]. 作物杂志, 2025, (3): 172–177 |
| [10] | 李虎, 黄秋要, 吴子帅, 刘广林, 陈传华, 罗群昌, 朱其南. 种植密度和施氮量对优质常规稻桂育12产量和米质的影响[J]. 作物杂志, 2025, (3): 195–201 |
| [11] | 陈虎, 高原, 孙家猛, 俞鹏, 肖洪武, 张海涛. 大麦典型硫氧还蛋白(TRX)基因家族鉴定与生物信息学分析[J]. 作物杂志, 2025, (2): 66–73 |
| [12] | 姬景红, 刘双全, 马星竹, 郝小雨, 郑雨, 赵月, 王晓军, 匡恩俊. 控释尿素对寒地水稻农艺性状、产量及氮肥利用率的影响[J]. 作物杂志, 2025, (2): 149–154 |
| [13] | 金丹丹, 隋世江, 陈玥, 李波, 曲航, 宫亮. 秸秆还田下氮肥减量对辽河平原水稻产量及氮素利用的影响[J]. 作物杂志, 2025, (2): 172–179 |
| [14] | 伍露, 张皓, 杨霏云, 郭尔静, 斯林林, 曹凯, 程陈. WOFOST模型对江淮地区水稻生长发育模拟的适应性评价[J]. 作物杂志, 2025, (2): 215–221 |
| [15] | 江素珍, 许超, 王中元, 郑沈, 陈建国, 朱捍华, 黄道友, 张泉, 朱奇宏. 海泡石和生物炭对水稻镉和砷吸收积累的影响[J]. 作物杂志, 2025, (2): 241–248 |
|
||