作物杂志,2021, 第6期: 36–45 doi: 10.16035/j.issn.1001-7283.2021.06.006
利用染色体片段置换系定位低温影响水稻萌芽期根长和芽长QTL
余镁霞1( ), 邓浩东1(
), 邓浩东1( ), 谭景艾1, 宋贵廷1, 吴光亮1, 陈利平1, 刘睿琦1, 邹安东1, 贺浩华1,2, 边建民1,2(
), 谭景艾1, 宋贵廷1, 吴光亮1, 陈利平1, 刘睿琦1, 邹安东1, 贺浩华1,2, 边建民1,2( )
)
- 1作物生理生态与遗传育种教育部/江西省重点实验室,330045,江西南昌
2江西省水稻高水平工程研究中心,330045,江西南昌
Detection of QTL for Root Length and Bud Length at Germination Stage in Low Temperature Using CSSLs in Rice (Oryza sativa)
Yu Meixia1( ), Deng Haodong1(
), Deng Haodong1( ), Tan Jing’ai1, Song Guiting1, Wu Guangliang1, Chen Liping1, Liu Ruiqi1, Zhou Andong1, He Haohua1,2, Bian Jianmin1,2(
), Tan Jing’ai1, Song Guiting1, Wu Guangliang1, Chen Liping1, Liu Ruiqi1, Zhou Andong1, He Haohua1,2, Bian Jianmin1,2( )
)
- 1Key Laboratory of Crop Physiology, Ecology and Genetic Breeding (Jiangxi Agricultural University), Ministry of Education/Jiangxi Province, Nanchang 330045, Jiangxi, China
2Jiangxi Super Rice Engineering Technology Research Center, Nanchang 330045, Jiangxi, China
摘要:
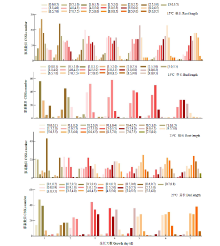
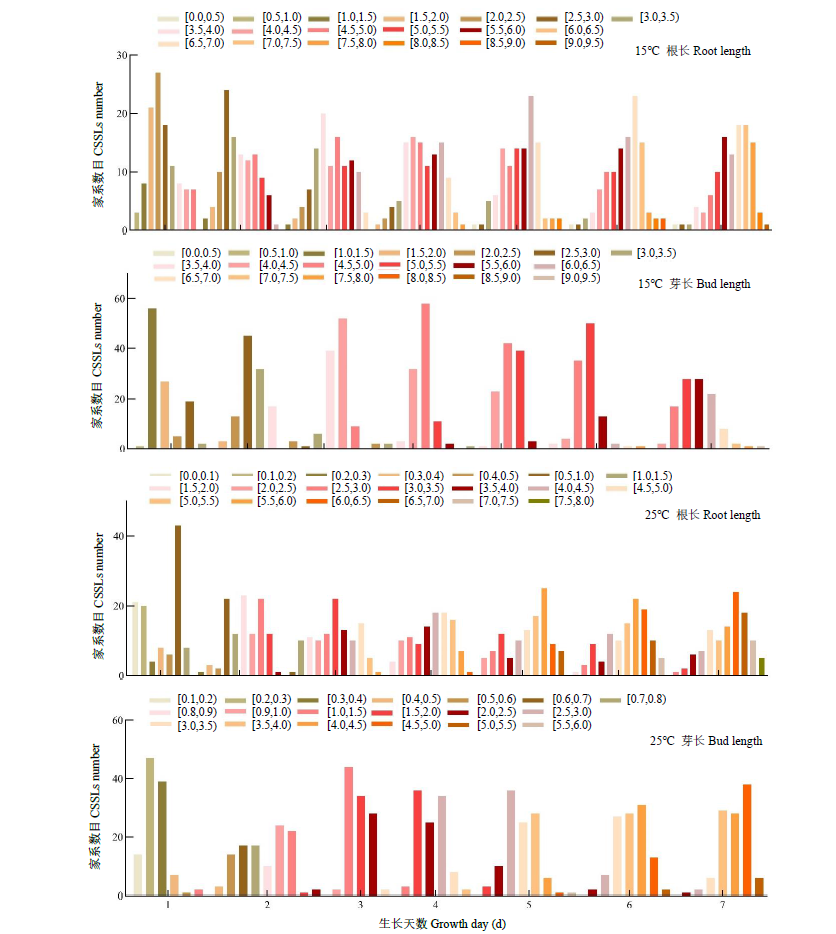
根和芽的正常生长和发育对保障水稻产量有重要作用。为探究在低温条件下影响幼芽期根长和芽长的QTL,以9311(受体)和日本晴(供体)染色体片段置换系群体为材料,将萌发种子置于15℃低温处理7d,然后在25℃下恢复生长7d,连续测量根长和芽长,以25℃下根长和芽长作为对照,定位低温条件下影响根长和芽长的QTL。15℃条件下,定位到9个根长QTL,贡献率为8.65%~24.35%,其中qRL15T7.2和qRL15T10.3在根生长不同时期被重复检测到;定位到9个影响芽长的QTL,贡献率为1.73%~28.89%,其中qBL15T6、qBL15T7、qBL15T9和qBL15T10被重复检测到。25℃条件下,定位到9个根长QTL,贡献率为1.73%~28.89%,qRL25T7、qRL25T9.3、qRL25T10和qRL25T12在根生长不同时期被重复检测到;定位到5个影响芽长QTL,贡献率为0.58%~38.26%,qBL25T3.3在芽生长不同时期被重复检测到。其中,第2、7、9和10号染色体上4个Bin标记区域存在既控制根长又控制芽长的QTL。通过比较发现,15℃下定位到的根长或芽长QTL与25℃定位到的根长或芽长QTL均不相同,表明低温条件下控制水稻幼芽期根和芽生长的遗传机制可能与正常温度下不同。
| [1] | Zhang Z Y, Li J J, Pan Y H, et al. Natural variation in CTB4a enhances rice adaptation to cold habitats. Nature Communications, 2017(8):14788. |
| [2] | 刘次桃, 王威, 毛毕刚, 等. 水稻耐低温逆境研究:分子生理机制及育种展望. 遗传, 2018, 40(3):171-185. |
| [3] |
Groot T T, Bodegom V P, Meijer H, et al. Gas Transport through the root-shoot transition zone of rice tillers. Plant and Soil, 2005, 277(1):107-116.
doi: 10.1007/s11104-005-0435-4 |
| [4] | 韩龙植, 乔永利, 曹桂兰, 等. 水稻生长早期耐冷性QTL分析. 中国水稻科学, 2005(2):122-126. |
| [5] | 杨洛淼, 王敬国, 刘化龙, 等. 寒地粳稻发芽期和芽期的耐冷性QTL定位. 作物杂志, 2014(6):44-51. |
| [6] |
Andaya V C, Tai T H. Fine mapping of the qCTS12 locus,a major QTL for seedling cold tolerance in rice. Theoretical and Applied Genetics, 2006, 113(3):467-475.
pmid: 16741739 |
| [7] |
Fujino K, Sekiguchi H. Origins of functional nucleotide polymorphisms in a major quantitative trait locus,qLTG3-1,controlling low-temperature germinability in rice. Plant Molecular Biology, 2011, 75(1/2):1-10.
doi: 10.1007/s11103-010-9697-1 |
| [8] |
Li J L, Pan Y H, Guo H F, et al. Fine mapping of QTL qCTB10-2 that confers cold tolerance at the booting stage in rice. Theoretical and Applied Genetics, 2018, 131(1):157.
doi: 10.1007/s00122-017-2992-3 |
| [9] |
Shi Y T, Yang S H. COLD1:a cold sensor in rice. Science China Life Sciences, 2015, 58(4):409-410.
doi: 10.1007/s11427-015-4831-6 |
| [10] |
Zhou L, Zeng Y W, Zheng W W, et al. Fine mapping a QTL qCTB7 for cold tolerance at the booting stage on rice chromosome 7 using a near-isogenic line. Theoretical and Applied Genetics, 2010, 121(5):895-905.
doi: 10.1007/s00122-010-1358-x pmid: 20512559 |
| [11] |
Steele K A, Price A H, Shashidhar H E, et al. Marker-assisted selection to introgress rice QTLs controlling root traits into an Indian upland rice variety. Theoretical and Applied Genetics, 2006, 112(2):208-221.
pmid: 16208503 |
| [12] |
Shimizu A, Kato K, Komatsu A, et al. Genetic analysis of root elongation induced by phosphorus deficiency in rice (Oryza sativa L.):fine QTL mapping and multivariate analysis of related traits. Theoretical and Applied Genetics, 2008, 117(6):987-996.
doi: 10.1007/s00122-008-0838-8 |
| [13] |
Mitsuhiro O, Wataru T, Takeshi E, et al. Fine-mapping of qRL6.1,a major QTL for root length of rice seedlings grown under a wide range of NH4+ concentrations in hydroponic conditions. Theoretical and Applied Genetics, 2010, 121(3):535-547.
doi: 10.1007/s00122-010-1328-3 pmid: 20390245 |
| [14] |
Wang H M, Xu X M, Zhan X D, et al. Identification of qRL7,a major quantitative trait locus associated with rice root length in hydroponic conditions. Breeding Science, 2013, 63(3):267-274.
doi: 10.1270/jsbbs.63.267 |
| [15] | 徐晓明, 张迎信, 王会民, 等. 一个水稻根长QTL qRL4的分离鉴定. 中国水稻科学, 2016, 30(4):363-370. |
| [16] | 赵春芳, 张亚东, 陈涛, 等. 低磷胁迫下水稻苗期根长性状的QTL定位. 华北农学报, 2013, 28(6):6-10. |
| [17] |
Redona E D, Mackill D J. Mapping quantitative trait loci for seedling vigor in rice using RFLPs. Theoretical and Applied Genetics, 1996, 92(3/4):395-402.
doi: 10.1007/BF00223685 |
| [18] |
Fukuda A, Terao T. QTLs for shoot length and Chlorophyll content of rice seedlings grown under low-temperature conditions,using a cross between Indica and Japonica cultivars. Plant Production Science, 2015, 18(2):128-136.
doi: 10.1626/pps.18.128 |
| [19] |
Zhang Z H, Yu S B, Yu T, et al. Mapping quantitative trait loci (QTLs) for seedling-vigor using recombinant inbred lines of rice (Oryza sativa L.). Field Crops Research, 2004, 91(2):161-170.
doi: 10.1016/j.fcr.2004.06.004 |
| [20] | 班超, 张晓玲, 穆平. 水稻根系性状QTL的整合、分类和真实性分析. 中国农学通报, 2009, 25(19):20-25. |
| [21] | Meng L, Li H H, Zhang L Y, et al. QTL IciMapping:Integrated software for genetic linkage map construction and quantitative trait locus mapping in bi-parental populations. The Crop Journal, 2015(3):269-283. |
| [22] |
Li H H, Ye G Y, Wang J K. A modified algorithm for the improvement of composite interval mapping. Genetics, 2007, 175(1):361-374.
doi: 10.1534/genetics.106.066811 |
| [23] |
McCouch S R. Committee on gene symbolization,nomenclature and linkage,rice genetics cooperative. Rice, 2008, 1(1):72-84.
doi: 10.1007/s12284-008-9004-9 |
| [24] | 王存虎, 刘东, 许锐能, 等. 大豆叶柄角的QTL定位分析. 作物学报, 2020, 46(1):9-19. |
| [25] | 陈利华, 万杉. 不同温度条件下水稻种子活力QTL的定位分析. 武汉植物学研究, 2005, 23(2):125-130. |
| [26] | 代贵金, 华泽田, 陈温福, 等. 杂交粳稻、常规粳稻、旱稻及籼稻根系特征比较. 沈阳农业大学学报, 2008, 39(5):515-519. |
| [27] | 蔡昆争, 骆世明, 段舜山. 水稻群体根系特征与地上部生长发育和产量的关系. 华南农业大学学报, 2005, 26(2):1-4. |
| [28] |
Fujino K, Sekiguchi H, Sato T, et al. Mapping of quantitative trait loci controlling low-temperature germinability in rice (Oryza sativa L.). Theoretical and Applied Genetics, 2004, 108(5):794-799.
pmid: 14624339 |
| [29] |
Miura K, Lin S Y, Yano M, et al. Mapping quantitative trait loci controlling low-temperature germinability in rice (Oryza sativa L.). Breeding Science, 2001, 51(4):293-299.
doi: 10.1270/jsbbs.51.293 |
| [30] |
Courtois B, Shen L, Petalcorin W, et al. Locating QTLs controlling constitutive root traits in the rice population IAC 165×Co39. Euphytica, 2003, 134(3):335-345.
doi: 10.1023/B:EUPH.0000004987.88718.d6 |
| [31] |
Zheng H G, Babu R C, Pathan M S, et al. Quantitative trait loci for root-penetration ability and root thickness in rice:comparison of genetic backgrounds. Genome, 2000, 43(1):53-61.
pmid: 10701113 |
| [32] | 徐吉臣, 李晶昭, 郑先武, 等. 苗期水稻根部性状的QTL定位. 遗传学报, 2001, 28(5):433-439. |
| [33] |
Uga Y, Okuno K, Yano M. QTLs underlying natural variation in stele and xylem structures of rice root. Breeding Science, 2008, 58(1):7-14.
doi: 10.1270/jsbbs.58.7 |
| [34] |
Ikeda H, Kamoshita A, Manabe T. Genetic analysis of rooting ability of transplanted rice (Oryza sativa L.) under different water conditions. Journal of Experimental Botany, 2007, 58(2):309-318
pmid: 17075079 |
| [35] |
Li W X, Zhao H J, Pang W Q, et al. Seed-specific silencing of OsMRP5 reduces seed phytic acid and weight in rice. Transgenic Research, 2014, 23(4):585-599.
doi: 10.1007/s11248-014-9792-1 |
| [36] |
Yukihiro I, Fumiko K, Kazuma H, et al. Fatty acid elongase is required for shoot development in rice. Plant Journal, 2011, 66(4):680-688.
doi: 10.1111/j.1365-313X.2011.04530.x |
| [37] |
Zhao Y, Jiang C H, Rehman R, et al. Genetic analysis of roots and shoots in rice seedling by association mapping. Genes and Genomics, 2019, 41(1):95-105.
doi: 10.1007/s13258-018-0741-x pmid: 30242741 |
| [1] | 李旭, 付立东, 王宇, 隋鑫, 任海, 吕小红, 马畅, 杜萌, 毛艇. DEP1与NRT1.1B基因的遗传互作对水稻氮素利用的影响[J]. 作物杂志, 2021, (6): 22–27 |
| [2] | 张俊, 邓艾兴, 尚子吟, 唐志伟, 严圣吉, 张卫建. 秸秆还田下水稻丰产与甲烷减排的技术模式[J]. 作物杂志, 2021, (6): 230–235 |
| [3] | 苏代群, 陈亮, 李锋, 武琦, 白君杰, 邹德堂, 王敬国, 刘化龙, 郑洪亮. 利用高密度遗传图谱发掘水稻抽穗期新位点[J]. 作物杂志, 2021, (6): 58–61 |
| [4] | 石楠, 高志强, 胡海燕, 陈崇怡, 文双雅. 杂交稻有序机抛增密减肥处理对产量及肥料偏生产力的影响[J]. 作物杂志, 2021, (5): 128–133 |
| [5] | 唐志强, 张丽颖, 何娜, 马作斌, 赵明珠, 王昌华, 郑文静, 银永安, 王辉. 机械旱直播对水稻生育进程、光合特性及产量的影响[J]. 作物杂志, 2021, (5): 87–94 |
| [6] | 吴可, 谢慧敏, 刘文奇, 莫并茂, 韦国良, 陆献, 李壮林, 邓森霞, 韦善清, 梁和, 江立庚. 氮、磷、钾肥对南方双季稻区水稻产量及产量构成因子的影响[J]. 作物杂志, 2021, (4): 178–183 |
| [7] | 凌晨, 刘洪, 杨哲, 黄展权, 陈孟强, 饶得花, 徐振江. 双季稻栽培对水稻DUS测试标准品种数量性状表达的影响[J]. 作物杂志, 2021, (4): 18–25 |
| [8] | 张全芳, 姜明松, 陈峰, 朱文银, 周学标, 杨连群, 徐建第. 山东省水稻品种(系)的遗传多样性分析[J]. 作物杂志, 2021, (4): 26–31 |
| [9] | 王国骄, 宋鹏, 杨振中, 张文忠. 秸秆还田对水稻光合物质生产特征、稻米品质和土壤养分的影响[J]. 作物杂志, 2021, (4): 67–72 |
| [10] | 薛菁芳, 蔡永盛, 陈书强. 节水灌溉栽培模式对稻米品质和淀粉RVA谱的影响[J]. 作物杂志, 2021, (4): 86–92 |
| [11] | 杨磊, 金延迪, 刘侯俊. 铁、镉及其互作对水稻光合原初反应的影响[J]. 作物杂志, 2021, (4): 144–151 |
| [12] | 孟祥宇, 冉成, 刘宝龙, 赵哲萱, 白晶晶, 耿艳秋. 秸秆还田配施氮肥对东北黑土稻区土壤养分及水稻产量的影响[J]. 作物杂志, 2021, (3): 167–172 |
| [13] | 邢愿, 贺中华. 基于水分亏缺指数的贵州省水稻干旱特征分析[J]. 作物杂志, 2021, (2): 191–199 |
| [14] | 付景, 尹海庆, 王亚, 杨文博, 张珍, 白涛, 王越涛, 王付华, 王生轩. 不同追氮模式对河南沿黄稻区粳稻根系生长和产量的影响[J]. 作物杂志, 2021, (2): 77–86 |
| [15] | 周江, 谢宜章, 向平安. 湖南主要大田作物系统投入产出的能值分析[J]. 作物杂志, 2021, (1): 175–181 |
|
||