作物杂志,2022, 第3期: 55–62 doi: 10.16035/j.issn.1001-7283.2022.03.008
谷子黄叶色突变体ylm-1的精细定位与功能分析
秦娜( ), 朱灿灿, 代书桃, 宋迎辉, 李君霞(
), 朱灿灿, 代书桃, 宋迎辉, 李君霞( ), 王春义
), 王春义
- 河南省农业科学院粮食作物研究所,450002,河南郑州
Fine Mapping and Functional Analysis of Yellow Leaf Mutant ylm-1 in Foxtail Millet
Qin Na( ), Zhu Cancan, Dai Shutao, Song Yinghui, Li Junxia(
), Zhu Cancan, Dai Shutao, Song Yinghui, Li Junxia( ), Wang Chunyi
), Wang Chunyi
- Cereal Crops Institute, Henan Academy of Agricultural Sciences, Zhengzhou 450002, Henan, China
摘要:
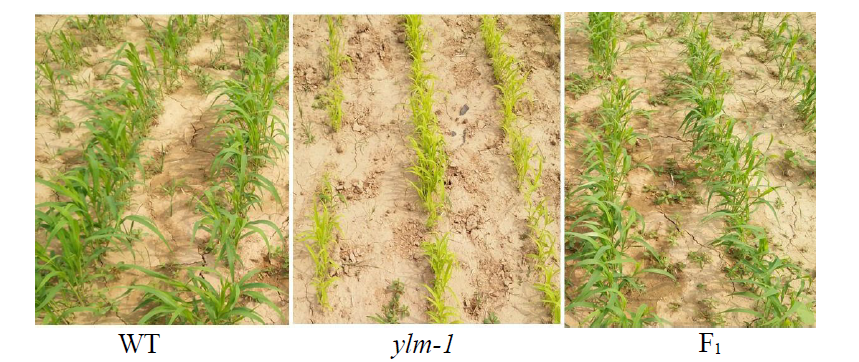
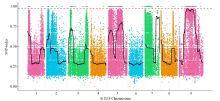
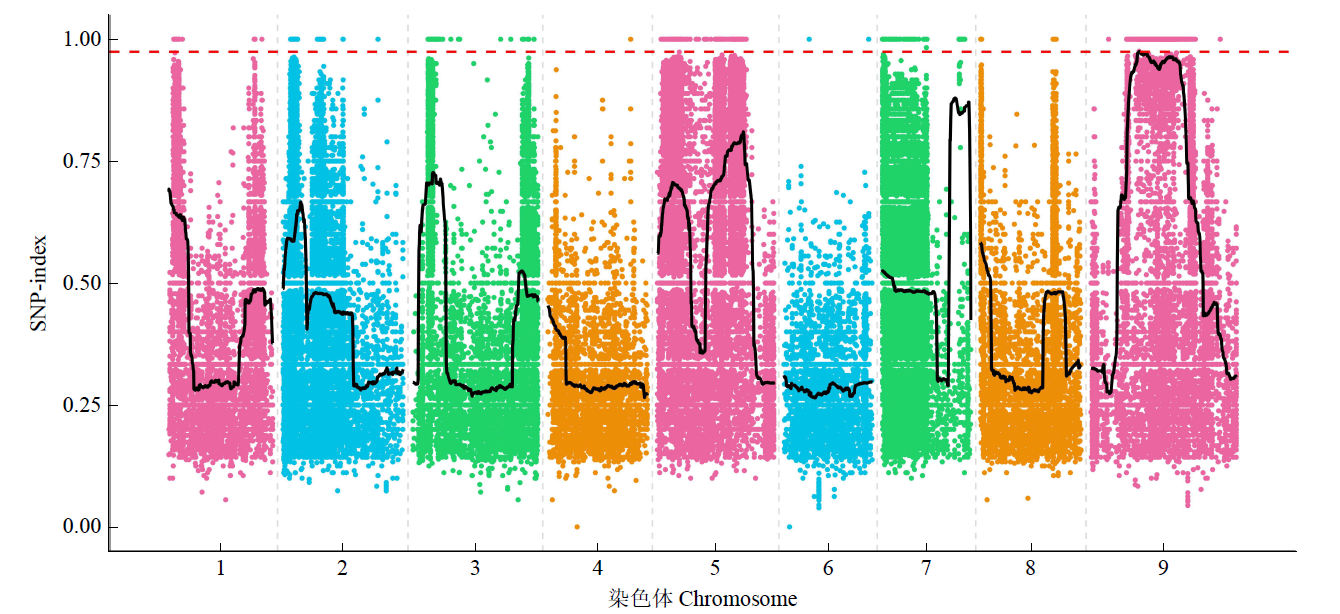
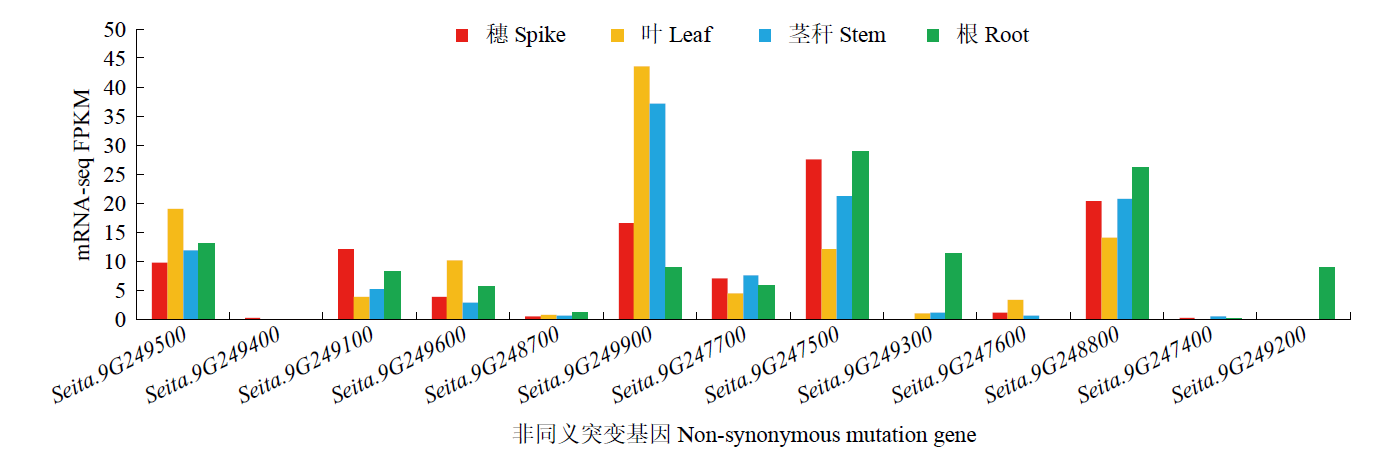
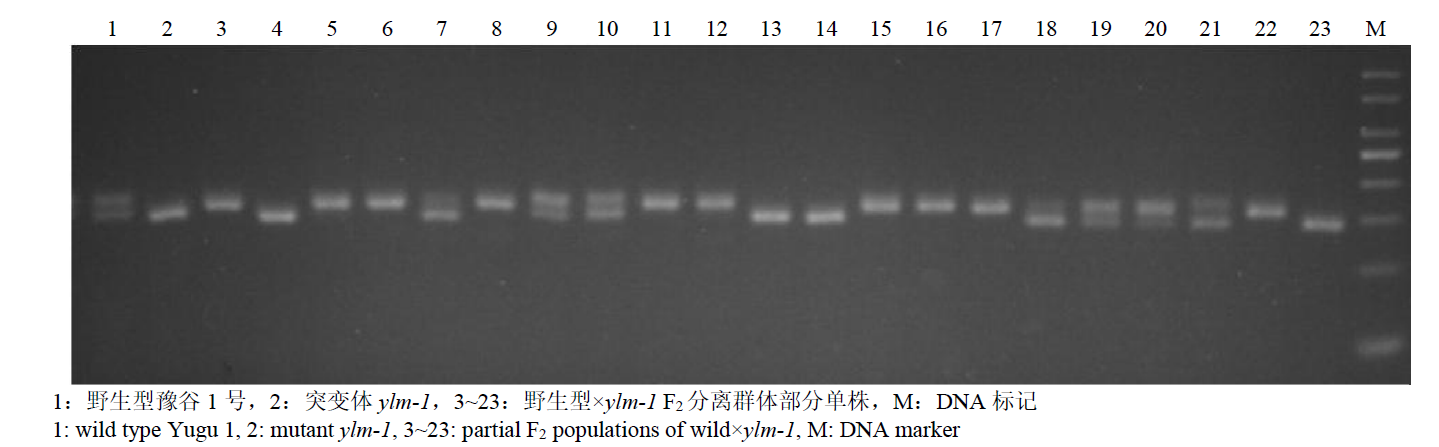
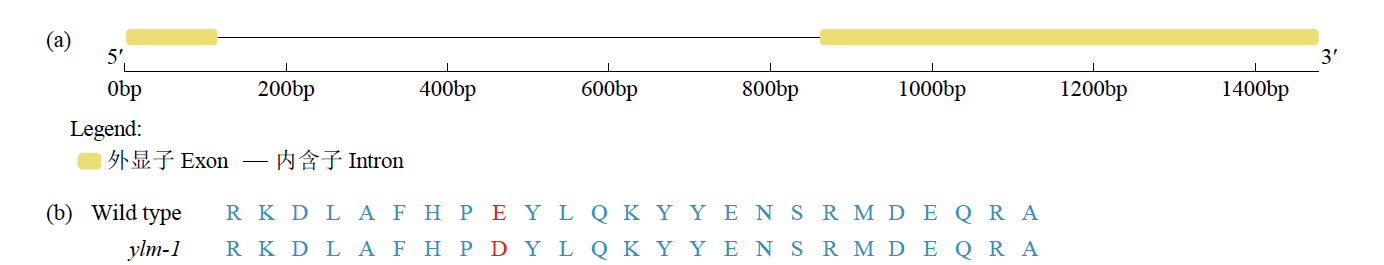
谷子叶色突变体在研究C4光能利用效率和叶绿素代谢机制方面具有重要作用。为探究谷子叶色突变的分子机制,利用甲基磺酸乙酯(EMS)处理谷子品种豫谷1号,获得1个稳定遗传的黄叶色突变体ylm-1。通过表型和遗传分析及遗传背景检测,同时利用突变位点图谱(MutMap)技术对突变基因进行精细定位。结果表明,ylm-1整个生育期叶色均为淡黄色;ylm-1与野生型遗传背景相同且受1对隐性核基因控制;MutMap精细定位到第9号染色体关联区间内共13个非同义突变基因,其中,Seita.9G249900是一个编码与红叶绿素代谢还原酶(RCCR)相关的基因,该基因位于第9号染色体19 937 988与19 940 620bp之间,含有2个外显子和1个内含子,在第2外显子361bp处发生A/T碱基突变,导致1个谷氨酸(E)变成天冬氨酸(D)。根据突变碱基设计了dCAPS标记,进一步验证了ylm-1候选基因突变位点。黄叶色突变体ylm-1的鉴定与基因功能分析将为该基因的有效利用、功能验证及作用机制研究奠定理论与技术基础。
| [1] |
Zhao S L, Long W H, Wang Y H, et al. A rice White-stripe leaf (wsl3) mutant lacking an HD domain-containing protein affects chlorophyll biosynthesis and chloroplast development. Journal of Plant Biology, 2016, 59(3):282-292.
doi: 10.1007/s12374-016-0459-8 |
| [2] |
Rooijen R V, Kruijer W, Boesten R, et al. Natural variation of YELLOW SEEDLING 1 affects photosynthetic acclimation of Arabidopsis thaliana. Nature Communications, 2017, 8(1):1424-1432.
doi: 10.1038/s41467-017-01408-4 |
| [3] |
Wang N, Liu Z Y, Zhang Y, et al. Identification and fine mapping of a stay-green gene (Brnye1) in pakchoi (Brassica campestris L. ssp. chinensis). Theoretical and Applied Genetics, 2018, 131(3):673-684.
doi: 10.1007/s00122-017-3028-8 pmid: 29209732 |
| [4] | 邹金财, 张维林, 夏明辉, 等. 水稻阶段性温敏白化转绿突变体stgra254的特征和基因定位. 华北农学报, 2017, 32(3):1-6. |
| [5] |
Junqueira N E G, Ortiz S B, Leal C M V, et al. Anatomy and ultrastructure of embryonic leaves of the C4 species Setaria viridis. Annals of Botany, 2018, 121(6):1163-1172.
doi: 10.1093/aob/mcx217 pmid: 29415162 |
| [6] |
Jia G Q, Huang X H, Zhi H, et al. A haplotype map of genomic variations and genome wide association studies of agronomic traits in foxtail millet (Setaria italica). Nature Genetics, 2013, 45(8):957-961.
doi: 10.1038/ng.2673 |
| [7] | 贾冠清, 刁现民. 谷子(Setaria italica (L. ) P. Beauv.)作为功能基因组研究模式植物的发展现状及趋势. 生命科学, 2017, 29(3):292-301. |
| [8] | 胡彬华, 王平, 杜安平, 等. 水稻淡黄叶突变体pyl3的鉴定和基因定位. 核农学报, 2021, 35(12):2696-2703. |
| [9] | Wang Y, He F, Liu J L, et al. Mapped clone and functional analysis of leaf color gene Ygl7 in a rice hybrid (Oryza sativa L. ssp. indica). PLoS ONE, 2014, 9(6):1-11. |
| [10] | Guan H Y, Xu X B, He C M, et al. Fine mapping and candidate gene analysis of the leaf-color gene ygl-1 in maize. PLoS ONE, 2016, 11(4):1-19. |
| [11] | 李传宗. 谷子苗期黄叶性状的生理基础及候选基因鉴定. 北京: 中国农业科学院, 2020. |
| [12] |
Li W, Tang S, Zhang S, et al. Gene mapping and functional analysis of the novel leaf color gene SiYGL1 in foxtail millet (Setaria italica (L.) P. Beauv). Physiologia Plantarum, 2016, 157(1):24-37.
doi: 10.1111/ppl.12405 |
| [13] |
Tang C J, Tang S, Zhang S, et al. SiSTL1encoding a large subunit of ribonudeotide reductase,is crucial for plant growth,chloroplast biogenesis,and cell cycle progression in Setaria italica. Journal of Experimental Botany, 2019, 70(4):1167-1182.
doi: 10.1093/jxb/ery429 |
| [14] | Zhang S, Tang S, Tang C J, et al. SiSTL2 is required for cell cycle,leaf organ development,chloroplast biogenesis, and has effects on C4 photosynthesis in Setaria italica (L.) P. Beauv. Frontiers in Plant Science, 2018, 9(1):1103-1113. |
| [15] |
Abe A, Kosugi S, Yoshida K, et al. Genome sequencing reveals agronomically important loci in rice using MutMap. Nature Biotechnology, 2012, 30(2):174-178.
doi: 10.1038/nbt.2095 |
| [16] |
Uchida N, Sakamoto T, Kurata T, et al. Identification of EMS induced causal mutations in a non-reference Arabidopsis thaliana accession by whole genome sequencing. Plant and Cell Physiology, 2011, 52(4):716-722.
doi: 10.1093/pcp/pcr029 |
| [17] |
Takagi H, Abe A, Yoshida K, et al. QTL-seq:rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant Journal, 2013, 74(1):174-183.
doi: 10.1111/tpj.12105 |
| [18] |
Tran Q, Bui N, Kappel C, et al. Mapping-by-sequencing via MutMap identifies a mutation in ZmCLE 7 underlying fasciation in a newly developed EMS mutant population in an elite tropical maize inbred. Genes, 2020, 11(3):281.
doi: 10.3390/genes11030281 |
| [19] | 王秋兰, 王智兰, 韩芳, 等. 谷子条纹叶突变体wsl2的鉴定及候选基因分析. 华北农学报, 2020, 35(1):214-221. |
| [20] | 李君霞, 秦娜, 朱灿灿, 等. 谷子黄叶色突变体光合特性研究. 核农学报, 2021, 35(9):1964-1970. |
| [21] | You Q, Zhang L W, Yi X, et al. SIFGD:Setaria italica functional genomics database. Molecular Plant, 2015, 8(6):967-970. |
| [22] |
Liu X P, Yang C, Han F Q, et al. Genetics and fine mapping of a yellow-green leaf gene (ygl-1) in cabbage (Brassica oleracea var. capitata L.). Molecular Breeding, 2016, 36:1-8.
doi: 10.1007/s11032-015-0425-z |
| [23] | James G V, Patel V, Nordström K J, et al. User guide for Mapping by sequencing in Arabidopsis. Genome Biology, 2013, 14(6):61-73. |
| [24] | 李洋洋, 薛冰, 周倩, 等. 黄瓜叶色黄化突变基因yl-2的鉴定. 中国蔬菜, 2020(7):30-37. |
| [25] | 陈竹锋, 严维, 王娜, 等. 利用改进的MutMap方法克隆水稻雄性不育基因. 遗传, 2014, 36(1):85-93. |
| [26] | 袁金红, 李俊华, 袁娇娇, 等. 基于全基因组测序的MutMap方法在正向遗传学研究中的应用. 遗传, 2017, 39(12):1168-1177. |
| [27] |
郭广君, 刁卫平, 刘金兵, 等. 辣椒抗黄瓜花叶病毒病研究进展. 华北农学报, 2014, 29(S1):77-84.
pmid: 19374909 |
|
Sugishima M, Kitamori Y, Noguchi M, et al. Crystal structure of red chlorophyll catabolite reductase:enlargement of the ferredoxin-dependent bilin reductase family. Journal of Molecular Biology, 2009, 389(2):376-387.
doi: 10.1016/j.jmb.2009.04.017 pmid: 19374909 |
|
| [28] |
Pruzinská A, Anders I, Aubry S, et al. In vivo participation of red chlorophyll catabolite reductase in chlorophyll breakdown. Plant Cell, 2007, 19(1):369-387.
pmid: 17237353 |
| [29] |
Xie Z K, Wu S D, Chen J Y, et al. The C‑terminal cysteine-rich motif of NYE1/SGR1 is indispensable for its function in chlorophyll degradation in Arabidopsis. Plant Molecular Biology, 2019, 101(1):257-268.
doi: 10.1007/s11103-019-00902-1 |
| [30] |
Schelbert S, Aubry S, Burla B, et al. Pheophytin pheophorbide hydrolase (pheophytinase) is involved in chlorophyll breakdown during leaf senescence in Arabidopsis. Plant Cell, 2009, 21(3):767-785.
doi: 10.1105/tpc.108.064089 pmid: 19304936 |
| [31] |
Chen Y, Shimoda Y, Yokono M, et al. Mg-dechelatase is involved in the formation of photosystem II but not in chlorophyll degradation in Chlamydomonas reinhardtii. The Plant Journal, 2009, 97(1):1022-1031.
doi: 10.1111/tpj.14174 |
| [32] |
Hauenstein M, Christ B, Das A, et al. A role for TIC 55 as a hydroxylase of phyllobilins,the products of chlorophyll breakdown during plant senescence. Plant Cell, 2016, 28(1):2510-2527.
doi: 10.1105/tpc.16.00630 |
| [33] |
Li Z, Wu S, Chen J, et al. NYEs/SGRs-mediated chlorophyll degradation is critical for detoxification during seed maturation in Arabidopsis. Plant Journal, 2017, 92(1):650-661.
doi: 10.1111/tpj.13710 |
| [34] |
Pruzinská A, Anders I, Aubry S, et al. In vivo participation of red chlorophyll catabolite reductase in chlorophyll breakdown. Plant Cell, 2007, 19(1):369-387.
pmid: 17237353 |
| [35] |
Fukasawa A, Suzuki Y, Terai H, et al. Effects of postharvest ethanol vapor treatment on activities and gene expression of chlorophyll catabolic enzymes in broccoli florets. Postharvest Biology and Technology, 2010, 55(2):97-102.
doi: 10.1016/j.postharvbio.2009.08.010 |
| [36] |
Gomez-Lobato M E, Civello P M, Martinez G A. Effects of ethylene,cytokinin and physical treatments on BoPaO gene expression of harvested broccoli. Journal of the Science of Food and Agriculture, 2012, 92(1):151-158.
doi: 10.1002/jsfa.4555 pmid: 21732385 |
| [1] | 郑思怡, 杨晔, 宋远辉, 花芹, 林泉祥, 张海涛, 程治军. 水稻甜质胚乳突变体m5788的鉴定及基因定位[J]. 作物杂志, 2022, (4): 14–21 |
| [2] | 马珂, 冯雷, 赵夏童, 张丽光, 原向阳, 董淑琦, 郭平毅, 宋喜娥. 播距和播量对张杂谷10号生长特性及产量的影响[J]. 作物杂志, 2022, (4): 172–178 |
| [3] | 吕建珍, 任莹, 王宏勇, 张庭军, 马建萍, 赵凯. 264份谷子主要育成品种(系)表型多样性综合评价[J]. 作物杂志, 2022, (4): 22–31 |
| [4] | 李秉华, 王贵启, 师志刚, 刘小民, 许贤, 赵铂锤, 程汝宏. 谷子和杂草对氰氟草酯的敏感性[J]. 作物杂志, 2022, (4): 262–266 |
| [5] | 刘攀锋, 秦杰, 郝爽楠, 王丹立, 杨武德, 冯美臣, 宋晓彦. 硒肥浓度、施用时期和施肥方式对不同谷子品种产量和籽粒硒含量的影响[J]. 作物杂志, 2022, (2): 182–188 |
| [6] | 郭永新, 周浩, 孙鹏, 王雅情, 马珂, 李晓瑞, 董淑琦, 郭平毅, 原向阳. 种植方式对不同地区张杂谷10号抗倒伏特性及产量的影响[J]. 作物杂志, 2022, (2): 195–202 |
| [7] | 赵利蓉, 马珂, 张丽光, 汤沙, 原向阳, 刁现民. 不同生态区谷子品种农艺性状和品质分析[J]. 作物杂志, 2022, (2): 44–53 |
| [8] | 卢平, 康庆芳, 赵孟瑶, 张凤洁, 武强强, 马芳芳, 王宇珅, 韩渊怀, 王兴春, 李雪垠. 谷子miR169家族及其靶基因的鉴定与功能分析[J]. 作物杂志, 2022, (2): 54–63 |
| [9] | 李会霞, 刘红, 王玉文, 田岗, 刘鑫, 郑植尹. 谷子抗除草剂杂交种去除假杂种技术研究[J]. 作物杂志, 2021, (6): 72–77 |
| [10] | 刘鑫, 李会霞, 田岗, 王玉文, 刘红, 曹晋军, 成锴, 王振华, 刘永忠, 李万星. 全生育期水分控制对谷子生长发育及品质的影响[J]. 作物杂志, 2021, (5): 181–186 |
| [11] | 王雨婷, 苗兴芬, 王帝. 萌发期耐莠去津谷子种质资源筛选及评价[J]. 作物杂志, 2021, (5): 194–204 |
| [12] | 高鹏, 郭美俊, 杨雪芳, 董淑琦, 温银元, 郭平毅, 原向阳. 谷子和玉米叶片光合荧光参数对烟嘧磺隆胁迫的响应差异[J]. 作物杂志, 2021, (3): 70–77 |
| [13] | 申洁, 王玉国, 郭平毅, 原向阳. 腐植酸对干旱胁迫下谷子幼苗叶片抗坏血酸-谷胱甘肽循环的影响[J]. 作物杂志, 2021, (2): 173–177 |
| [14] | 贾苏卿, 禾璐, 杜艳伟. 不同耕作方式对旱区春谷根系发育、产量及水分利用效率的影响[J]. 作物杂志, 2020, (5): 194–198 |
| [15] | 杨永青, 高芳芳, 马亚君, 陈鑫, 张杰. 山西省旱作农业区不同施肥处理对谷子产量、品质及经济效益的影响[J]. 作物杂志, 2020, (4): 195–201 |
|
||