作物杂志,2022, 第2期: 54–63 doi: 10.16035/j.issn.1001-7283.2022.02.008
谷子miR169家族及其靶基因的鉴定与功能分析
卢平1( ), 康庆芳1, 赵孟瑶1, 张凤洁2, 武强强2, 马芳芳2, 王宇珅2, 韩渊怀2, 王兴春1, 李雪垠1,*(
), 康庆芳1, 赵孟瑶1, 张凤洁2, 武强强2, 马芳芳2, 王宇珅2, 韩渊怀2, 王兴春1, 李雪垠1,*( )
)
- 1山西农业大学生命科学学院,030801,山西晋中
2山西农业大学农学院,030801,山西晋中
Identification and Functional Analysis of miR169 Family and Its Target Genes in Setaria italica
Lu Ping1( ), Kang Qingfang1, Zhao Mengyao1, Zhang Fengjie2, Wu Qiangqiang2, Ma Fangfang2, Wang Yushen2, Han Yuanhuai2, Wang Xingchun1, Li Xueyin1,*(
), Kang Qingfang1, Zhao Mengyao1, Zhang Fengjie2, Wu Qiangqiang2, Ma Fangfang2, Wang Yushen2, Han Yuanhuai2, Wang Xingchun1, Li Xueyin1,*( )
)
- 1College of Life Sciences, Shanxi Agricultural University, Jinzhong 030801, Shanxi, China
2College of Agriculture, Shanxi Agricultural University, Jinzhong 030801, Shanxi, China
摘要:
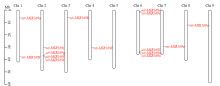
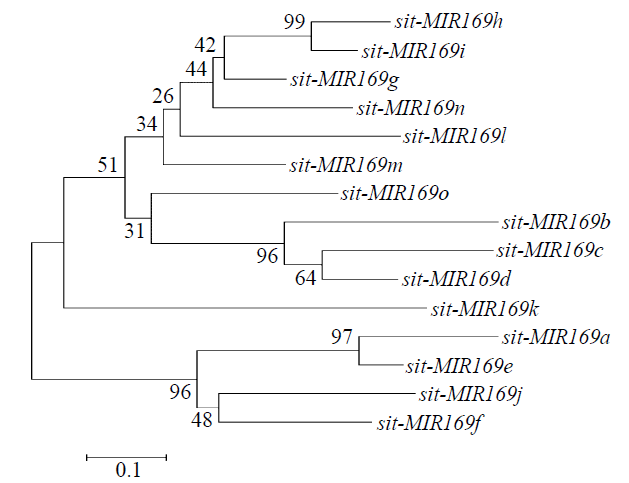
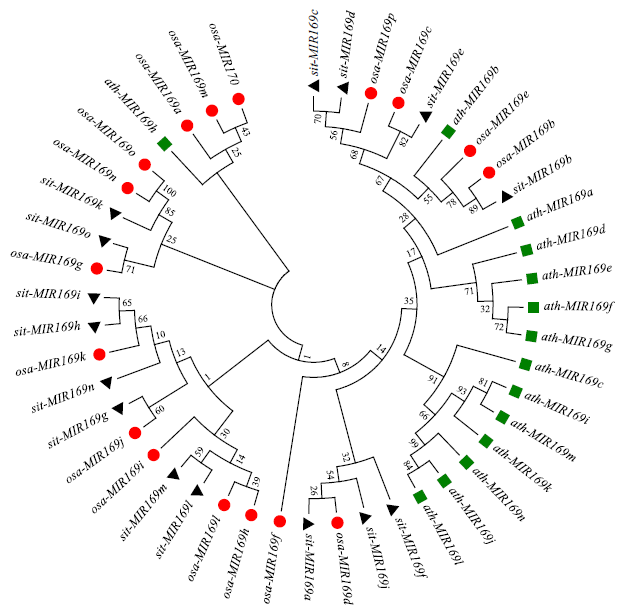
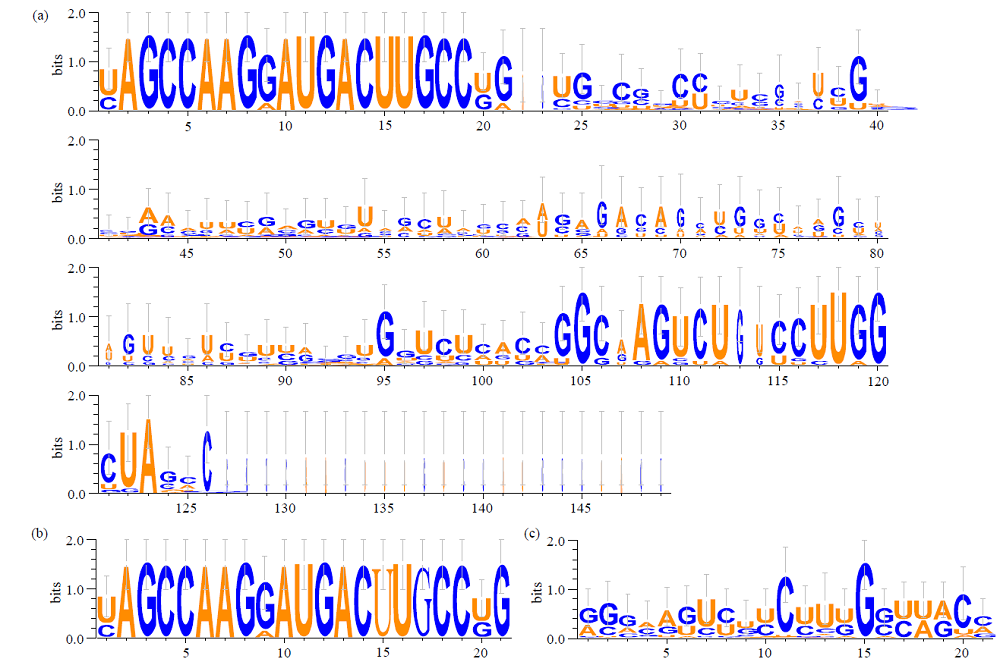
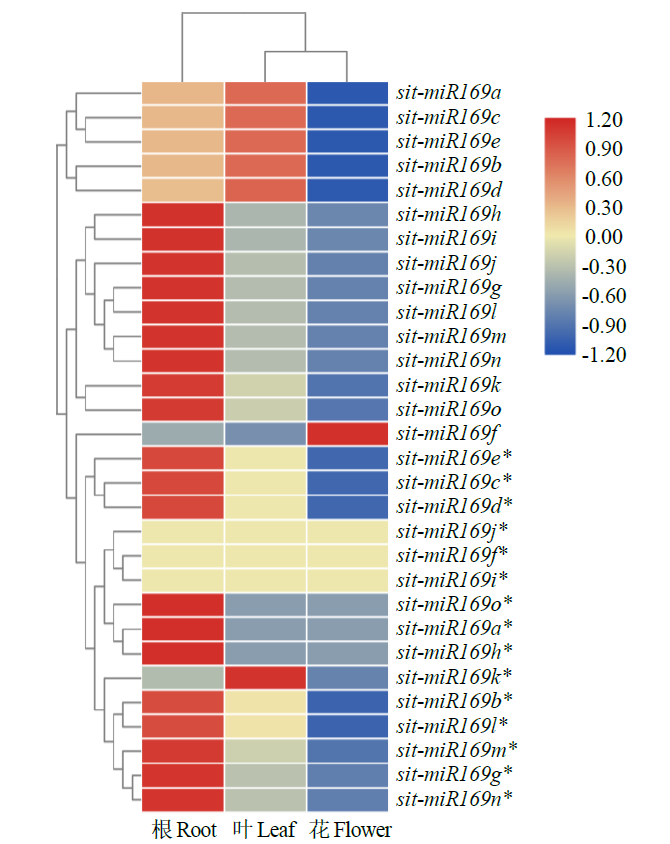
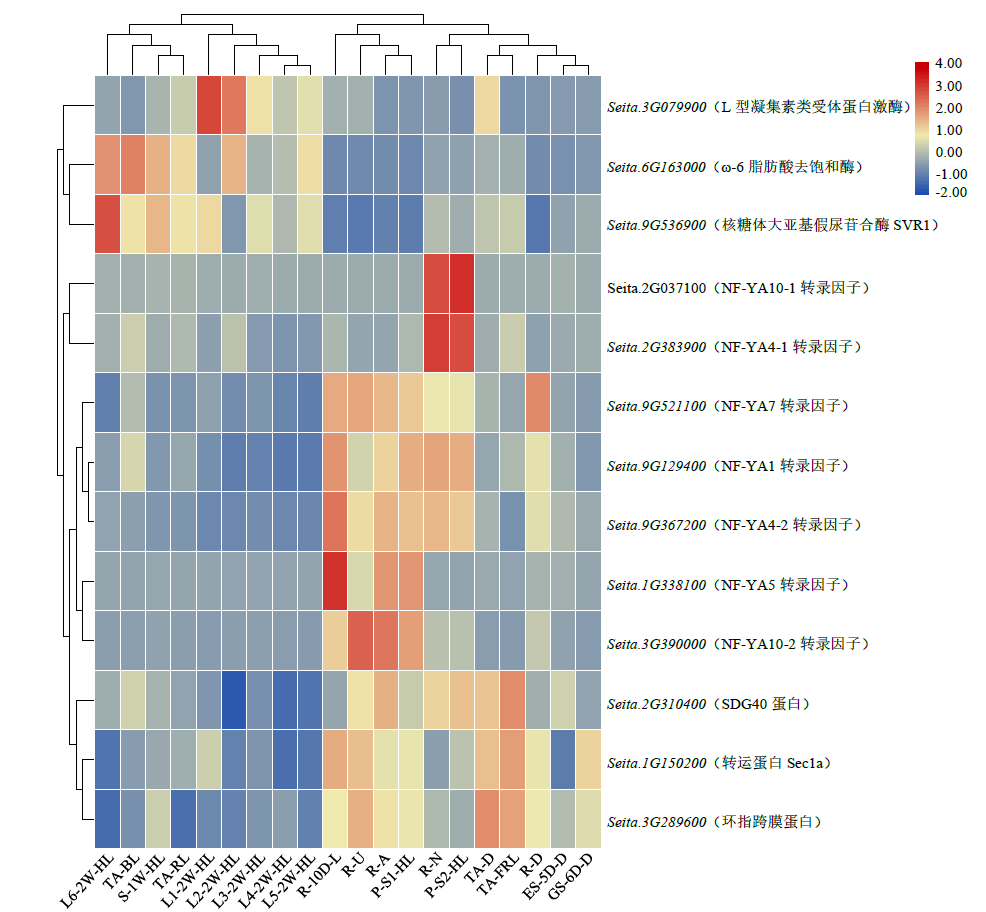
microRNA广泛参与植物中多个生长发育过程,miR169家族是最保守的miRNA家族之一。对谷子miR169(sit-miR169)家族进行生物信息学分析,包括染色体分布、系统进化、碱基保守性、二级结构和sit-miR169及其靶基因的表达。鉴定了15个sit-miR169,分布于7条染色体上。系统进化分析显示,sit-miR169家族前体sit-MIR169大致可以分为2簇,部分sit-MIR169成员与水稻osa-MIR169家族成员亲缘关系较近。sit-miR169的前体均可形成稳定的二级茎环结构,成熟体均产生于前体的5′端臂上,且成熟序列的碱基保守性很强。sit-miR169在花、叶和根中均有表达,具有明显组织表达特异性。sit-miR169的靶基因主要是NF-YA转录因子基因,此外还有L型凝集素类受体蛋白激酶、ω-6脂肪酸去饱和酶、转运蛋白Sec1a、SDG40蛋白、环指跨膜蛋白和核糖体大亚基假尿苷合酶SVR1基因等。miR169通过调控靶基因,尤其是NF-YA的表达量来影响植物的生长发育和对环境的应答。谷子NF-YA基因主要在根和穗中表达,表现出明显的组织表达特异性。此外,sit-miR169靶基因的表达受环境诱导,如干旱条件能进一步诱导Seita.9G521100基因在根中的表达。
| [1] | 刁现民. 中国谷子产业与产业技术体系. 北京: 中国农业科学技术出版社, 2011. |
| [2] |
Diao X M, Schnable J, Bennetzen J L, et al. Initiation of Setaria as a model plant. Frontiers of Agricultural Science and Engineering, 2014, 1(1):16.
doi: 10.15302/J-FASE-2014011 |
| [3] |
Yang Z R, Zhang H S, Li X K, et al. A mini foxtail millet with an Arabidopsis-like life cycle as a C4 model system. Nature Plants, 2020, 6(9):1167-1178.
doi: 10.1038/s41477-020-0747-7 |
| [4] |
Bennetzen J L, Schmutz J, Wang H, et al. Reference genome sequence of the model plant Setaria. Nature Biotechnology, 2012, 30(6):555-561.
doi: 10.1038/nbt.2196 pmid: 22580951 |
| [5] |
Zhang G Y, Liu X, Quan Z W, et al. Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nature Biotechnology, 2012, 30(6):549-554.
doi: 10.1038/nbt.2195 |
| [6] |
杨丽娟, 李世访, 卢美光. miRNA 在植物病原调控方面的研究进展. 生物技术通报, 2020, 36(1):101-109.
doi: 10.13560/j.cnki.biotech.bull.1985.2019-1045 |
| [7] | 郁佳雯, 郁佳雯, 韩荣鹏, 等. microRNA 在植物生长发育中的研究进展. 分子植物育种, 2020, 18(5):1496-1504. |
| [8] |
张翠桔, 莫蓓莘, 陈雪梅, 等. 植物 miRNA 作用方式的分子机制研究进展. 生物技术通报, 2020, 36(7):1-14.
doi: 10.13560/j.cnki.biotech.bull.1985.2020-0262 |
| [9] | Wang Y Q, Li L, Tang S, et al. Combined small RNA and degradome sequencing to identify miRNAs and their targets in response to drought in foxtail millet. BMC Genetics, 2016, 17(1):1-16 |
| [10] |
Xu M Y, Zhang L, Li W W, et al. Stress-induced early flowering is mediated by miR169 in Arabidopsis thaliana. Journal of Experimental Botany, 2014, 65(1):89-101.
doi: 10.1093/jxb/ert353 |
| [11] | 赵胜利.Osa-miR169 调控水稻对稻瘟病菌免疫机理的研究. 成都:四川农业大学, 2017. |
| [12] |
Zhao B T, Ge L F, Liang R Q, et al. Members of miR-169 family are induced by high salinity and transiently inhibit the NF-YA transcription factor. BMC Molecular Biology, 2009, 10(1):1-10.
doi: 10.1186/1471-2199-10-1 |
| [13] |
Luan M D, Xu M Y, Lu Y M, et al. Expression of zma-miR169 miRNAs and their target ZmNF-YA genes in response to abiotic stress in maize leaves. Gene, 2015, 555(2):178-185.
doi: 10.1016/j.gene.2014.11.001 |
| [14] |
Ni Z Y, Hu Z, Jiang Q Y, et al. GmNFYA3,a target gene of miR169,is a positive regulator of plant tolerance to drought stress. Plant Molecular Biology, 2013, 82(1/2):113-129.
doi: 10.1007/s11103-013-0040-5 |
| [15] | 尹海龙.大豆 miR169d 的表达分析及功能验证. 长春:吉林农业大学, 2013. |
| [16] | 孙广鑫, 栾雨时, 崔娟娟. 番茄中与致病密切相关 miRNA 的挖掘及特性分析. 遗传, 2013, 36(1):69-76. |
| [17] | 董云, 王毅, 靳丰蔚, 等. 油菜 Bna-miR169d 基因的分离与过表达初步分析. 西北农业学报, 2016, 25(12):1809-1815. |
| [18] | 刘志祥, 曾超珍, 谭晓风. 杨树 MIR169 基因家族分子进化分析. 遗传, 2013, 35(11):1307-1316. |
| [19] | 方辉, 曲俊杰, 孙嘉曼, 等. 葡萄 miR169 及其靶基因的生物信息学分析. 南方农业学报, 2017, 48(8):1329-1334. |
| [20] | Donati G, Gatta R, Dolfini D, et al. An NF-Y-dependent switch of positive and negative histone methyl marks on CCAAT promoters. PLoS ONE, 2008, 3(4):e2066. |
| [21] | Nelson D E, Repetti P P, Adams T R, et al. Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proceedings of the National Academy of Sciences, 2007, 104(42):16450-16455. |
| [22] | Sinha S, Maity S N, Lu J F, et al. Recombinant rat CBF-C,the third subunit of CBF/NFY,allows formation of a protein-DNA complex with CBF-A and CBF-B and with yeast HAP2 and HAP3. Proceedings of the National Academy of Sciences, 1995, 92(5):1624-1628. |
| [23] |
Li W X, Oono Y, Zhu J H, et al. The Arabidopsis NFYA 5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. The Plant Cell, 2008, 20(8):2238-2251.
doi: 10.1105/tpc.108.059444 |
| [24] | Guo Z L, Kuang Z, Wang Y, et al. PmiREN:a comprehensive encyclopedia of plant miRNAs. Nucleic Acids Research, 2020, 48(D1):1114-1121. |
| [25] |
Chen C J, Chen H, Zhang Y, et al. TBtools-an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 2020, 13(8):1194-1202.
doi: 10.1016/j.molp.2020.06.009 |
| [26] |
Crooks G E, Hon G, Chandonia J M, et al. WebLogo:a sequence logo generator. Genome Research, 2004, 14(6):1188-1190.
doi: 10.1101/gr.849004 pmid: 15173120 |
| [27] | Dai X B, Zhuang Z H, Zhao P X. psRNATarget:a plant small RNA target analysis server (2017 release). Nucleic Acids Research, 2018, 46(W1):49-54. |
| [28] |
Zhang Z H, Teotia S, Tang J H, et al. Perspectives on microRNAs and phased small interfering RNAs in maize (Zea mays L.):functions and big impact on agronomic traits enhancement. Plants, 2019, 8(6):1-17.
doi: 10.3390/plants8010001 |
| [29] |
Devos K M, Wang Z M, Beales J, et al. Comparative genetic maps of foxtail millet (Setaria italica) and rice (Oryza sativa). Theoretical and Applied Genetics, 1998, 96(1):63-68.
doi: 10.1007/s001220050709 |
| [30] | 徐妙云, 朱佳旭, 张敏, 等. 植物miR169/NF-YA调控模块研究进展. 遗传, 2016, 38(8):700-706. |
| [31] | 戴晓峰, 肖玲, 武玉花, 等. 植物脂肪酸去饱和酶及其编码基因研究进展. 植物学报, 2007, 24(1):105-113. |
| [1] | 刘攀锋, 秦杰, 郝爽楠, 王丹立, 杨武德, 冯美臣, 宋晓彦. 硒肥浓度、施用时期和施肥方式对不同谷子品种产量和籽粒硒含量的影响[J]. 作物杂志, 2022, (2): 182–188 |
| [2] | 郭永新, 周浩, 孙鹏, 王雅情, 马珂, 李晓瑞, 董淑琦, 郭平毅, 原向阳. 种植方式对不同地区张杂谷10号抗倒伏特性及产量的影响[J]. 作物杂志, 2022, (2): 195–202 |
| [3] | 赵利蓉, 马珂, 张丽光, 汤沙, 原向阳, 刁现民. 不同生态区谷子品种农艺性状和品质分析[J]. 作物杂志, 2022, (2): 44–53 |
| [4] | 尹桂芳, 段迎, 杨晓琳, 蔡苏云, 王艳青, 卢文洁, 孙道旺, 贺润丽, 王莉花. 苦荞FtC4H基因克隆与生物信息学分析[J]. 作物杂志, 2022, (1): 77–83 |
| [5] | 李会霞, 刘红, 王玉文, 田岗, 刘鑫, 郑植尹. 谷子抗除草剂杂交种去除假杂种技术研究[J]. 作物杂志, 2021, (6): 72–77 |
| [6] | 刘鑫, 李会霞, 田岗, 王玉文, 刘红, 曹晋军, 成锴, 王振华, 刘永忠, 李万星. 全生育期水分控制对谷子生长发育及品质的影响[J]. 作物杂志, 2021, (5): 181–186 |
| [7] | 王雨婷, 苗兴芬, 王帝. 萌发期耐莠去津谷子种质资源筛选及评价[J]. 作物杂志, 2021, (5): 194–204 |
| [8] | 高鹏, 郭美俊, 杨雪芳, 董淑琦, 温银元, 郭平毅, 原向阳. 谷子和玉米叶片光合荧光参数对烟嘧磺隆胁迫的响应差异[J]. 作物杂志, 2021, (3): 70–77 |
| [9] | 申洁, 王玉国, 郭平毅, 原向阳. 腐植酸对干旱胁迫下谷子幼苗叶片抗坏血酸-谷胱甘肽循环的影响[J]. 作物杂志, 2021, (2): 173–177 |
| [10] | 贾苏卿, 禾璐, 杜艳伟. 不同耕作方式对旱区春谷根系发育、产量及水分利用效率的影响[J]. 作物杂志, 2020, (5): 194–198 |
| [11] | 杨永青, 高芳芳, 马亚君, 陈鑫, 张杰. 山西省旱作农业区不同施肥处理对谷子产量、品质及经济效益的影响[J]. 作物杂志, 2020, (4): 195–201 |
| [12] | 赵训超,徐晶宇,盖胜男,魏玉磊,许晓萱,丁冬,刘梦,张今杰,邵文静. 高粱硬脂酰-ACP脱氢酶基因(SbSAD)家族鉴定及不同发育阶段表达分析[J]. 作物杂志, 2020, (2): 20–27 |
| [13] | 赵丽娟,袁红梅,赵丽伟,郭文栋,李志江,李祥羽,马金丰,李延东,宋维富,杨雪峰,刘东军. 谷子矮秆突变体d93090的表型变异及其对赤霉素的敏感性分析[J]. 作物杂志, 2019, (6): 27–32 |
| [14] | 宋健,晓宁,王海岗,陈凌,王君杰,刘思辰,乔治军. SiASRs家族基因的鉴定及表达分析[J]. 作物杂志, 2019, (6): 33–42 |
| [15] | 张笛,苗兴芬,王雨婷. 100份谷子品种资源萌发期耐盐性评价及耐盐品种筛选[J]. 作物杂志, 2019, (6): 43–49 |
|
||