作物杂志,2023, 第1期: 20–29 doi: 10.16035/j.issn.1001-7283.2023.01.004
主要禾谷类作物DGAT基因家族比较分析
孟亚轩( ), 姚旭航, 孙颖琦, 赵心月, 王凤霞, 瓮巧云, 刘颖慧(
), 姚旭航, 孙颖琦, 赵心月, 王凤霞, 瓮巧云, 刘颖慧( )
)
- 河北北方学院农林科技学院,075000,河北张家口
Identification and Bioinformatics Analysis of DGAT Gene Family in Cereal Crops
Meng Yaxuan( ), Yao Xuhang, Sun Yingqi, Zhao Xinyue, Wang Fengxia, Weng Qiaoyun, Liu Yinghui(
), Yao Xuhang, Sun Yingqi, Zhao Xinyue, Wang Fengxia, Weng Qiaoyun, Liu Yinghui( )
)
- College of Agriculture and Forestry Science and Technology, Hebei North University, Zhangjiakou 075000, Hebei, China
摘要:
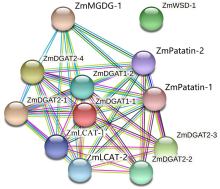
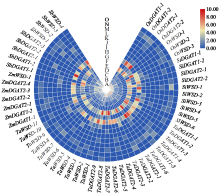
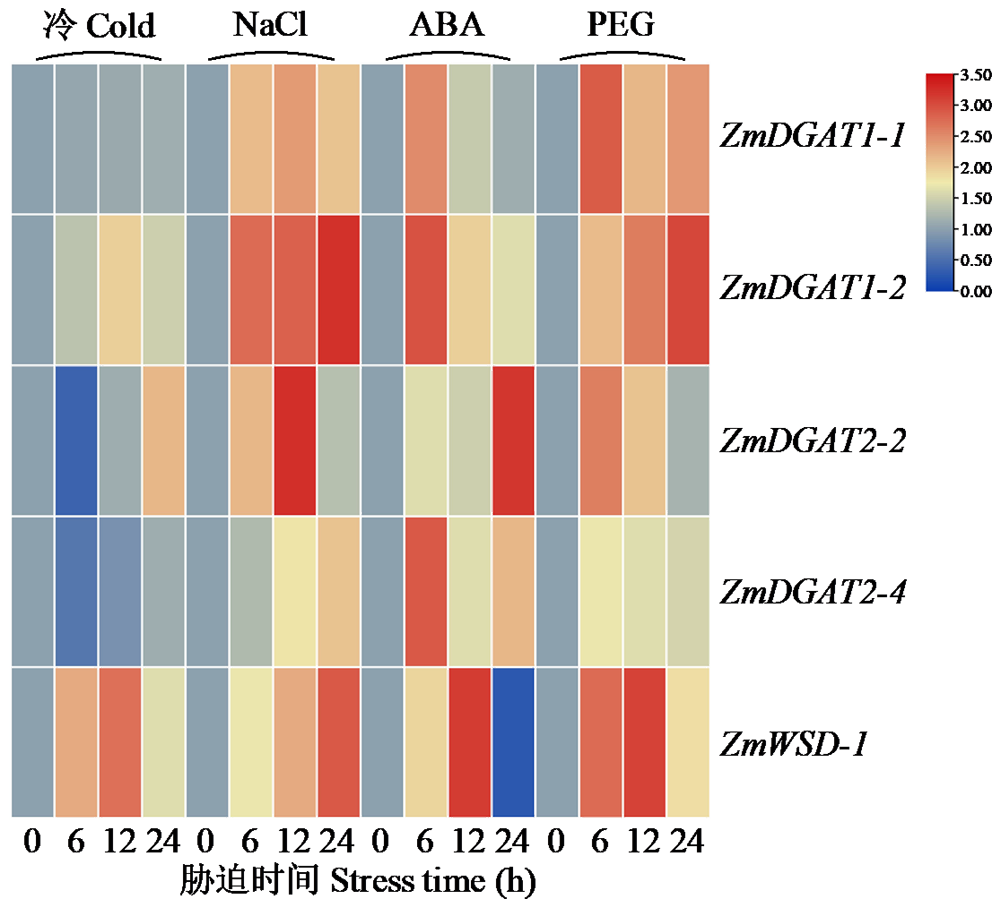
二酰甘油酰基转移酶(DGAT)是三酰甘油合成的限速酶,在植物油脂合成中发挥重要作用。为明确禾谷类作物DGAT基因家族的表达特征,利用已知DGAT成员序列对主要禾谷类作物进行全基因组扫描,分析其亚型差异及表达特点。结果表明,DGAT蛋白具有高脂肪、碱性和不稳定性。系统发育树中DGAT成员聚类为4个亚族,不同亚族成员间平行进化。DGAT基因结构具有组内保守性和组间多样性,基序存在多种类型,不同亚型成员具有特异的结构域组成。DGAT上游序列中,存在植株发育和胁迫响应等多种类型顺式作用元件,说明DGAT广泛参与不同生物学过程。转录组分析结果显示,DGAT广泛存在于不同组织中。在遭遇干旱和低温等不同非生物胁迫时,DGAT受到不同程度诱导,具有时空表达差异性。本研究可为禾谷类作物DGAT基因家族成员功能研究奠定基础。
| [1] |
Yang Y, Benning C. Functions of triacylglycerols during plant development and stress. Current Opinion in Biotechnology, 2018, 49:191-198.
doi: S0958-1669(17)30085-X pmid: 28987914 |
| [2] | Du Z Y, Benning C. Triacylglycerol Accumulation in photosynthetic cells in plants and algae. Subcellular Biochemistry, 2016, 86:179-205. |
| [3] |
Bhatt-Wessel B, Jordan T W, Miller J H, et al. Role of DGAT enzymes in triacylglycerol metabolism. Archives of Biochemistry and Biophysics, 2018, 655:1-11.
doi: S0003-9861(18)30190-5 pmid: 30077544 |
| [4] |
Chen G, Xu Y, Siloto R M P, et al. High-performance variants of plant diacylglycerol acyltransferase 1 generated by directed evolution provide insights into structure function. Plant Journal, 2017, 92(2):167-177.
doi: 10.1111/tpj.13652 |
| [5] |
Liu D, Ji H, Yang Z. Functional characterization of three novel genes encoding diacylglycerol acyltransferase (DGAT) from oil- rich tubers of cyperus esculentus. Plant and Cell Physiology. 2020, 61(1):118-129.
doi: 10.1093/pcp/pcz184 |
| [6] |
Zheng L, Shockey J, Guo F, et al. Discovery of a new mechanism for regulation of plant triacylglycerol metabolism: The peanut diacylglycerol acyltransferase-1 gene family transcriptome is highly enriched in alternative splicing variants. Journal of Plant Physiology, 2017, 219:62-70.
doi: S0176-1617(17)30242-0 pmid: 29031100 |
| [7] |
Zhou X R, Shrestha P, Yin F, et al. AtDGAT2 is a functional acyl-CoA:diacylglycerol acyltransferase and displays different acyl-CoA substrate preferences than AtDGAT1. FEBS Letters, 2013, 587(15):2371-2376.
doi: 10.1016/j.febslet.2013.06.003 |
| [8] |
Gao H, Gao Y, Zhang F, et al. Functional characterization of an novel acyl-CoA:diacylglycerol acyltransferase 3-3 (CsDGAT3-3) gene from camelina sativa. Plant Science, 2021, 303:110752.
doi: 10.1016/j.plantsci.2020.110752 |
| [9] |
Aymé L, Arragain S, Canonge M, et al. Arabidopsis thaliana DGAT 3 is a [2Fe-2S] protein involved in TAG biosynthesis. Scientific Reports, 2018, 8(1):17254.
doi: 10.1038/s41598-018-35545-7 |
| [10] | 陶芬芳. 甘蓝型油菜二酰甘油酰基转移酶(BnDGAT3)基因克隆与表达研究. 长沙:湖南农业大学, 2017. |
| [11] |
Rosli R, Chan P L, Chan K L, et al. In silico characterization and expression profiling of the diacylglycerol acyltransferase gene family (DGAT1,DGAT2,DGAT3 and WS/DGAT) from oil palm, Elaeis guineensis. Plant Science, 2018, 275:84-96.
doi: 10.1016/j.plantsci.2018.07.011 |
| [12] |
Yan B, Xu X, Gu Y, et al. Genome-wide characterization and expression profiling of diacylglycerol acyltransferase genes from maize. Genome, 2018, 61(10):735-743.
doi: 10.1139/gen-2018-0029 pmid: 30092654 |
| [13] | 闫博巍, 许晓萱, 谷英楠, 等. 玉米Ⅰ型二酰基转移酶基因(DGAT1)的生物信息学分析及其在非生物胁迫下的表达研究. 玉米科学, 2018, 26(1):20-28. |
| [14] |
Lu C, Hills M J. Arabidopsis mutants deficient in diacylglycerol acyltransferase display increased sensitivity to abscisic acid,sugars,and osmotic stress during germination and seedling development. Plant Physiology, 2002, 129(3):1352-1358.
doi: 10.1104/pp.006122 |
| [15] |
Siloto R M, Truksa M, Brownfield D, et al. Directed evolution of acyl-CoA:diacylglycerol acyltransferase:development and characterization of Brassica napus DGAT1 mutagenized libraries. Plant Physiology and Biochemistry, 2009, 47(6):456-461.
doi: 10.1016/j.plaphy.2008.12.019 |
| [16] |
Ortiz R, Geleta M, Gustafsson C, et al. Oil crops for the future. Current Opinion in Plant Biology, 2020, 56:181-189.
doi: S1369-5266(19)30118-9 pmid: 31982290 |
| [17] |
Singh Y, Sharma A, Singla A. Non-edible vegetable oil-based feedstocks capable of bio-lubricant production for automotive sector applications-a review. Environmental Science and Pollution Research, 2019, 26(15):14867-14882.
doi: 10.1007/s11356-019-05000-9 |
| [18] |
van Dijk E L, Jaszczyszyn Y, Naquin D, et al. The third revolution in sequencing technology. Trends in Genetics, 2018, 34(9):666-681.
doi: S0168-9525(18)30096-9 pmid: 29941292 |
| [19] | 郑玲, 单雷, 李新国, 等. 花生DGAT基因家族的生物信息学分析. 山东农业科学, 2018, 50(6):10-18. |
| [20] |
Chen C, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 2020, 13(8):1194-1202.
doi: S1674-2052(20)30187-8 pmid: 32585190 |
| [21] |
Liu Q, Siloto R M, Lehner R, et al. Acyl-CoA:diacylglycerol acyltransferase:molecular biology,biochemistry and biotechnology. Progress in Lipid Research, 2012, 51(4):350-377.
doi: 10.1016/j.plipres.2012.06.001 |
| [22] |
Cao H. Structure-function analysis of diacylglycerol acyltransferase sequences from 70 organisms. BMC Research Notes, 2011, 4:249.
doi: 10.1186/1756-0500-4-249 pmid: 21777418 |
| [23] |
Qiao X, Li Q, Yin H, et al. Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants. Genome Biology, 2019, 20(1):38.
doi: 10.1186/s13059-019-1650-2 pmid: 30791939 |
| [24] |
Xu C, Shanklin J. Triacylglycerol metabolism,function,and accumulation in plant vegetative tissues. Annual Review of Plant Biology, 2016, 67:179-206.
doi: 10.1146/annurev-arplant-043015-111641 |
| [1] | 周菲. 向日葵HaLACS7基因的生物信息学和表达分析[J]. 作物杂志, 2022, (3): 104–108 |
| [2] | 杨晓琳, 段迎, 蔡苏云, 贺润丽, 尹桂芳, 王艳青, 卢文洁, 孙道旺, 王莉花. 苦荞漆酶基因的克隆与生物信息学分析[J]. 作物杂志, 2022, (3): 73–79 |
| [3] | 尹桂芳, 段迎, 杨晓琳, 蔡苏云, 王艳青, 卢文洁, 孙道旺, 贺润丽, 王莉花. 苦荞FtC4H基因克隆与生物信息学分析[J]. 作物杂志, 2022, (1): 77–83 |
| [4] | 王志龙, 薛应红, 郝月茹, 刘宝玲, 苑丽霞, 薛金爱, 李润植. 亚麻荠油脂相关转录因子CsLEC2基因家族的鉴定及表达分析[J]. 作物杂志, 2020, (5): 23–32 |
| [5] | 徐园园, 赵鹏, 洪权春, 朱晓琴, 裴冬丽. 小麦转录因子基因TaMYB70的分离和表达分析[J]. 作物杂志, 2020, (4): 84–90 |
| [6] | 宋健,晓宁,王海岗,陈凌,王君杰,刘思辰,乔治军. SiASRs家族基因的鉴定及表达分析[J]. 作物杂志, 2019, (6): 33–42 |
| [7] | 岳琳祺,施卫萍,郭佳晖,郭平毅,郭杰. 谷子角质合成基因对干旱胁迫的响应[J]. 作物杂志, 2019, (4): 183–190 |
| [8] | 罗海斌, 蒋胜理, 黄诚梅, 曹辉庆, 邓智年, 吴凯朝, 徐林, 陆珍, 魏源文. 甘蔗ScHAK10基因克隆及表达分析[J]. 作物杂志, 2018, (4): 53–61 |
| [9] | 朱畇昊,爼梦航,苏秀红,董诚明,陈随清. 冬凌草HMGS基因的克隆与表达分析[J]. 作物杂志, 2016, (5): 25–30 |
| [10] | 李钰,郑文寅,冯春,王荣富,李娟. 非生物逆境胁迫下普通小麦烟农19幼苗FeSOD基因表达分析[J]. 作物杂志, 2016, (4): 75–79 |
| [11] | 袁红梅,郭文栋,赵丽娟,于莹,吴建忠,程莉莉,赵东升,康庆华,黄文功,姚玉波,宋喜霞,姜卫东,刘岩,马廷芬,吴广文,关凤芝. 亚麻糖基转移酶基因LuUGT72E1的克隆与表达分析[J]. 作物杂志, 2016, (4): 62–67 |
| [12] | 向鹏, 龙承波, 罗红丽. 水稻OsMED7基因的克隆及表达分析[J]. 作物杂志, 2013, (3): 21–24 |
| [13] | 刘允晶. 禾谷类作物耐盐细胞突变体筛选的研究进展[J]. 作物杂志, 1994, (6): 11–12 |
| [14] | 王海波. 细胞培养中细胞状态的调控[J]. 作物杂志, 1991, (3): 3–6 |
| [15] | 高立荣. 同源四倍体荞麦生物学特性观察[J]. 作物杂志, 1989, (3): 17–18 |
|
||