作物杂志,2025, 第2期: 66–73 doi: 10.16035/j.issn.1001-7283.2025.02.009
大麦典型硫氧还蛋白(TRX)基因家族鉴定与生物信息学分析
- 安徽农业大学农学院,230036,安徽合肥
Identification and Bioinformatics Analysis of the Typical Thioredoxin (TRX) Gene Family in Barley
Chen Hu( ), Gao Yuan, Sun Jiameng, Yu Peng, Xiao Hongwu, Zhang Haitao(
), Gao Yuan, Sun Jiameng, Yu Peng, Xiao Hongwu, Zhang Haitao( )
)
- College of Agronomy, Anhui Agricultural University, Hefei 230036, Anhui, China
摘要:
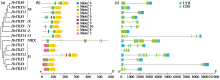
在栽培大麦MOREX全基因组水平下,利用TBtools、ExPASy、BUSCA、MEME、PlantCARE等在线工具对其染色体定位、理化性质、基因结构、保守结构域和启动子进行了预测分析。利用已发表的RNA-seq数据分析了典型TRX基因家族成员在大麦不同时期、不同组织部位的表达模式及其在干旱胁迫下的表达谱。结果表明,大麦典型TRX基因家族有15个成员,不均匀地分布在6条染色体上(除6H染色体)。启动子顺式作用元件预测到大麦典型TRX基因家族在生长发育、激素调控、非生物胁迫方面均发挥功能。15个典型TRX基因在种子、根、茎、叶和花等不同组织和不同时期均有表达。
| [1] |
Haas M, Schreiber M, Mascher M. Domestication and crop evolution of wheat and barley: Genes, genomics, and future directions. Journal of Integrative Plant Biology, 2019, 61(3):204-225.
doi: 10.1111/jipb.12737 |
| [2] | Geng L, Li M D, Zhang G D, et al. Barley: a potential cereal for producing healthy and functional foods. Food Quality and Safety, 2022, 6(2):142-154. |
| [3] | Sakellariou M, Mylona P V. New uses for traditional crops: the case of barley biofortification. Agronomy, 2020, 10(12):1964. |
| [4] | Lemaire S D, Miginiac-Maslow M. The thioredoxin superfamily in Chlamydomonas reinhardtii. Photosynthesis Research, 2004,82:203-220. |
| [5] |
Arnér E S J, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. European Journal of Biochemistry, 2000, 267(20):6102-6109.
doi: 10.1046/j.1432-1327.2000.01701.x pmid: 11012661 |
| [6] |
Liu J, Liu B, Feng D R, et al. Evidence for a role of chloroplastic m-type thioredoxins in the biogenesis of photosystem II in Arabidopsis. Plant Physiology, 2013, 163(4):1710-1728.
doi: 10.1104/pp.113.228353 pmid: 24151299 |
| [7] | Zhou J F, Song T Q, Zhou H W, et al. Genome-wide identification, characterization, evolution, and expression pattern analyses of the typical thioredoxin gene family in wheat (Triticum aestivum L.). Frontiers in Plant Science, 2022,13:1020584. |
| [8] | Jacquot J P, Vidal J, Gadal P, et al. Evidence for the existence of several enzyme-specific thioredoxins in plants. FEBS Letters, 1978, 96(2):243-246. |
| [9] | Wolosiuk R A, Buchanan B B. Thioredoxin and glutathione regulate photosynthesis in chloroplasts. Nature, 1977, 266(5602):565-567. |
| [10] |
Okegawa Y, Motohashi K. Chloroplastic thioredoxin m functions as a major regulator of Calvin cycle enzymes during photosynthesis in vivo. The Plant Journal, 2015, 84(5):900-913.
doi: 10.1111/tpj.13049 pmid: 26468055 |
| [11] |
Barajas-López J D, Serrato A J, Cazalis R, et al. Circadian regulation of chloroplastic f and m thioredoxins through control of the CCA 1 transcription factor. Journal of Experimental Botany, 2011, 62(6):2039-2051.
doi: 10.1093/jxb/erq394 pmid: 21196476 |
| [12] | Buchanan B B. The path to thioredoxin and redox regulation in chloroplasts. Annual Review of Plant Biology, 2016,67:1-24. |
| [13] | Gelhaye E, Rouhier N, Gérard J, et al. A specific form of thioredoxin h occurs in plant mitochondria and regulates the alternative oxidase. Proceedings of the National Academy of Sciences of the United States of America, 2004, 101(40):14545-14550. |
| [14] | Laloi C, Rayapuram N, Chartier Y, et al. Identification and characterization of a mitochondrial thioredoxin system in plants. Proceedings of the National Academy of Sciences of the United States of America, 2001, 98(24):14144-14149. |
| [15] | Collin V, Issakidis-Bourguet E, Marchand C, et al. The Arabidopsis plastidial thioredoxins: new functions and new insights into specificity. Journal of Biological Chemistry, 2003, 278(26):23747-23752. |
| [16] | Collin V, Lamkemeyer P, Miginiac-Maslow M, et al. Characterization of plastidial thioredoxins from Arabidopsis belonging to the new y-type. Plant Physiology, 2004, 136(4):4088-4095. |
| [17] | Arsova B, Hoja U, Wimmelbacher M, et al. Plastidial thioredoxin z interacts with two fructokinase-like proteins in a thiol- dependent manner: evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana. The Plant Cell, 2010, 22(5):1498-1515. |
| [18] |
Geigenberger P, Thormählen I, Daloso D M, et al. The unprecedented versatility of the plant thioredoxin system. Trends in Plant Science, 2017, 22(3):249-262.
doi: S1360-1385(16)30221-7 pmid: 28139457 |
| [19] |
Nikkanen L, Rintamäki E. Chloroplast thioredoxin systems dynamically regulate photosynthesis in plants. Biochemical Journal, 2019, 476(7):1159-1172.
doi: 10.1042/BCJ20180707 pmid: 30988137 |
| [20] |
Laughner B J, Sehnke P C, Ferl R J. A novel nuclear member of the thioredoxin superfamily. Plant Physiology, 1998, 118(3):987-996.
pmid: 9808743 |
| [21] |
Funato Y, Hayashi T, Irino Y, et al. Nucleoredoxin regulates glucose metabolism via phosphofructokinase 1. Biochemical and Biophysical Research Communications, 2013, 440(4):737-742.
doi: 10.1016/j.bbrc.2013.09.138 pmid: 24120946 |
| [22] | Chen C J, Chen H, Zhang Y, et al. TBtools:an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 2020, 13(8):1194-1202. |
| [23] |
Sun L J, Ren H Y, Liu R X, et al. An h-type thioredoxin functions in tobacco defense responses to two species of viruses and an abiotic oxidative stress. Molecular Plant-Microbe Interactions, 2010, 23(11):1470-1485.
doi: 10.1094/MPMI-01-10-0029 pmid: 20923353 |
| [24] |
Havelda Z, Várallyay É, Válóczi A, et al. Plant virus infection‐induced persistent host gene downregulation in systemically infected leaves. The Plant Journal, 2008, 55(2):278-288.
doi: 10.1111/j.1365-313X.2008.03501.x pmid: 18397378 |
| [25] | Valerio C, Costa A, Marri L, et al.Thioredoxin-regulated β-amylase (BAM1) triggers diurnal starch degradation in guard cells, and in mesophyll cells under osmotic stress. Journal of Experimental Botany, 2011, 62(2):545-555. |
| [26] | Moon H, Lee B, Choi G, et al. NDP kinase 2 interacts with two oxidative stress-activated MAPKs to regulate cellular redox state and enhances multiple stress tolerance in transgenic plants. Proceedings of the National Academy of Sciences of the United States of America, 2003, 100(1):358-363. |
| [27] |
Xie G, Kato H, Sasaki K, et al. A cold-induced thioredoxin h of rice, OsTrx23, negatively regulates kinase activities of OsMPK3 and OsMPK6 in vitro. FEBS Letters, 2009, 583(17):2734-2738.
doi: 10.1016/j.febslet.2009.07.057 pmid: 19665023 |
| [28] | Zhang S X, Yu Y, Song T Q, et al. Genome-wide identification of foxtail millet’s TRX family and a functional analysis of SiNRX1 in response to drought and salt stresses in transgenic Arabidopsis. Frontiers in Plant Science, 2022,13:946037. |
| [29] |
Nuruzzaman M, Sharoni A M, Satoh K, et al. The thioredoxin gene family in rice: Genome-wide identification and expression profiling under different biotic and abiotic treatments. Biochemical and Biophysical Research Communications, 2012, 423(2):417-423.
doi: 10.1016/j.bbrc.2012.05.142 pmid: 22683629 |
| [30] | Elasad M, Wei H L, Wang H T, et al. Genome-wide analysis and characterization of the TRX gene family in upland cotton. Tropical Plant Biology, 2018, 11(3/4):119-130. |
| [31] | Zhang J R, Zhao T, Yan F D, et al. Genome-wide identification and expression analysis of Thioredoxin (Trx) genes in seed development of vitis vinifera. Journal of Plant Growth Regulation, 2022, 41(7):3030-3045. |
| [32] | Li X, Su G J, Ntambiyukuri A, et al. Genome-wide identification and expression analysis of the AhTrx family genes in peanut. Biologia Plantarum, 2022, 66(1):112-122. |
| [33] | Bhurta R, Hurali D T, Tyagi S, et al. Genome-wide identification and expression analysis of the thioredoxin (trx) gene family reveals its role in leaf rust resistance in wheat (Triticum aestivum L.). Frontiers in Genetics, 2022,13:563. |
| [34] | Hägglund P, Björnberg O, Navrot N, et al. The barley grain thioredoxin system-an update. Frontiers in Plant Science, 2013,4:151. |
| [1] | 张俊, 蔡苏云, 徐子豪, 侯蕾, 贺润丽, 尹桂芳, 王莉花, 王艳青, 卢文洁, 孙道旺. 苦荞FtERF基因克隆、生物信息学及其表达分析[J]. 作物杂志, 2024, (2): 23–29 |
| [2] | 张倩, 任雯, 赵冰兵, 周秒依, 李韩帅, 刘亚, 杜何为. 玉米ZmMAPKKK21基因的克隆和生物信息学分析[J]. 作物杂志, 2024, (2): 30–39 |
| [3] | 张玉, 杨文静, 刘璇, 聂峰杰, 张丽, 石磊, 张国辉, 郭志乾, 巩檑. 马铃薯细胞壁蔗糖转化酶基因StCWIN1启动子克隆与表达及其在干旱胁迫下的作用分析[J]. 作物杂志, 2024, (2): 54–61 |
| [4] | 吕宝莲, 杨宇昕, 崔立操, 史峰, 马亮, 孔秀英, 张立超, 倪志勇. 小麦bHLH家族转录因子的鉴定及其在盐胁迫条件下的表达分析[J]. 作物杂志, 2024, (1): 65–72 |
| [5] | 毋莹, 胡蝶, 李婷, 段乾元, 韦宁宁, 张兴华, 徐淑兔, 薛吉全. 玉米WRKY转录因子IIc亚家族分析及其在干旱胁迫下的表达分析[J]. 作物杂志, 2024, (1): 80–89 |
| [6] | 夏雪, 蔡康锋, 刘磊, 宋秀娟, 汪军妹, 岳文浩. 大麦根形态和分子水平对低磷胁迫响应研究进展[J]. 作物杂志, 2023, (6): 11–16 |
| [7] | 赵锋, 包奇军, 潘永东, 柳小宁, 张华瑜, 牛小霞. 70份大麦种质资源遗传多样性评价[J]. 作物杂志, 2023, (6): 54–61 |
| [8] | 郜战宁, 杨永乾, 王树杰, 冯辉, 薛正刚. 143份大麦种质资源的综合评价[J]. 作物杂志, 2023, (5): 59–65 |
| [9] | 赵鹏鹏, 李鲁华, 任明见, 安畅, 洪鼎立, 李欣, 徐如宏. 小麦GzCIPK7-5B基因的生物信息学及表达分析[J]. 作物杂志, 2023, (4): 77–84 |
| [10] | 卢映吉, 杨晓梦, 普晓英, 李霞, 杨丽娥, 杨砚斌, 曾亚文. 不同季节播种和割苗对大麦优良品种农艺性状的影响[J]. 作物杂志, 2023, (3): 215–220 |
| [11] | 邱凯华, 方淑梅, 梁喜龙. 稻瘟病菌类SRRM1转录因子的功能分析[J]. 作物杂志, 2023, (3): 246–253 |
| [12] | 孟亚轩, 姚旭航, 孙颖琦, 赵心月, 王凤霞, 瓮巧云, 刘颖慧. 主要禾谷类作物DGAT基因家族比较分析[J]. 作物杂志, 2023, (1): 20–29 |
| [13] | 赵斌, 季昌好, 孙皓, 朱斌, 王瑞, 陈晓东. 多棱饲用大麦品系粮、草产量及品质的鉴定与综合评价[J]. 作物杂志, 2022, (6): 93–97 |
| [14] | 周菲. 向日葵HaLACS7基因的生物信息学和表达分析[J]. 作物杂志, 2022, (3): 104–108 |
| [15] | 杨晓琳, 段迎, 蔡苏云, 贺润丽, 尹桂芳, 王艳青, 卢文洁, 孙道旺, 王莉花. 苦荞漆酶基因的克隆与生物信息学分析[J]. 作物杂志, 2022, (3): 73–79 |
|