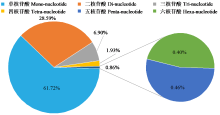
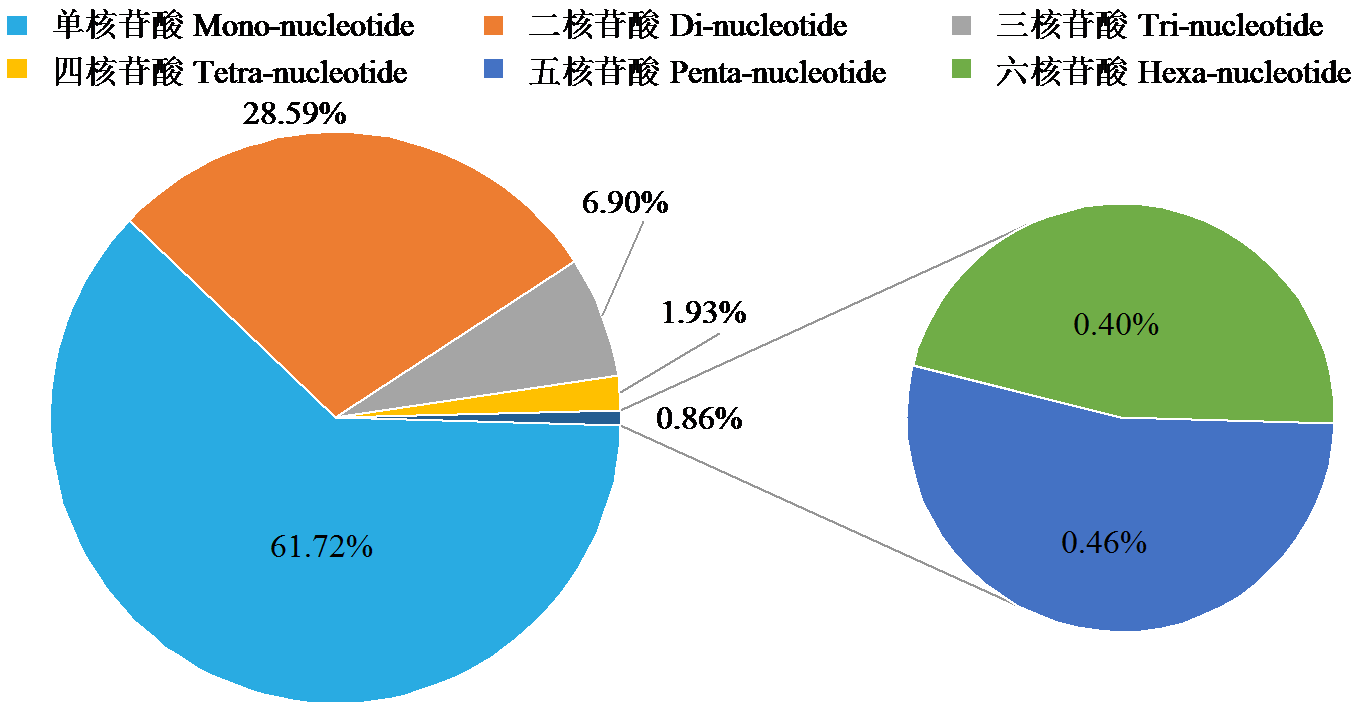
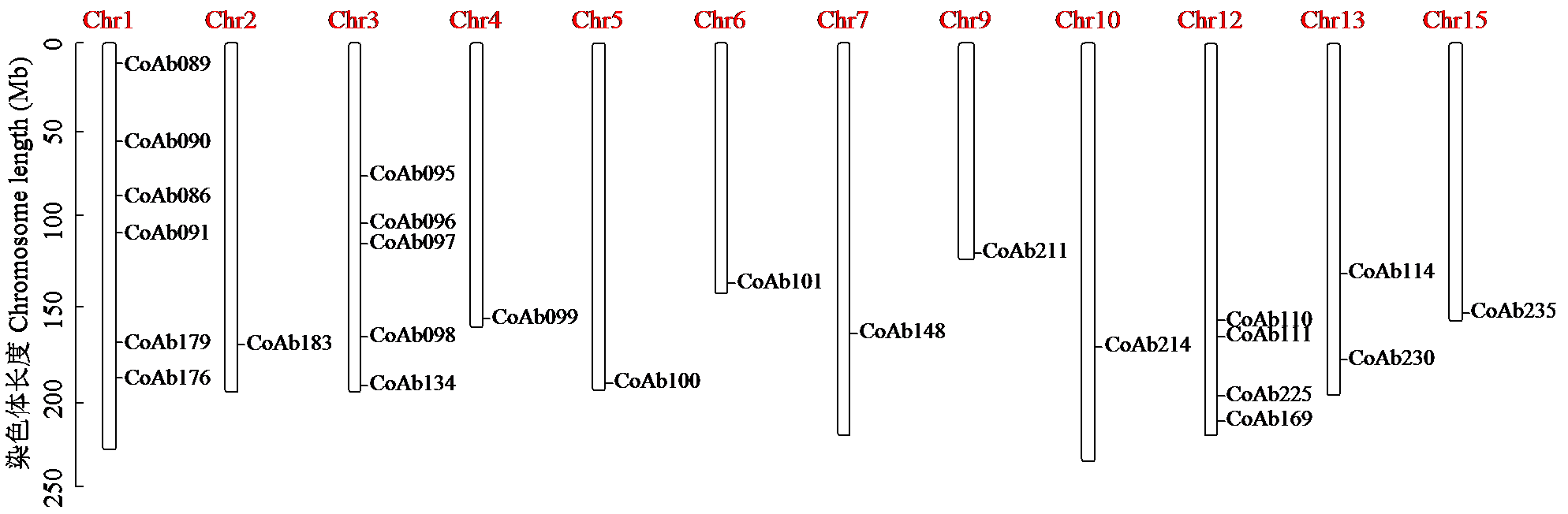
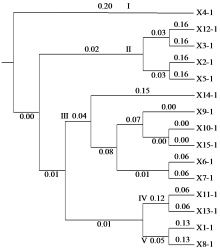
Crops ›› 2024, Vol. 40 ›› Issue (3): 23-31.doi: 10.16035/j.issn.1001-7283.2024.03.004
Previous Articles Next Articles
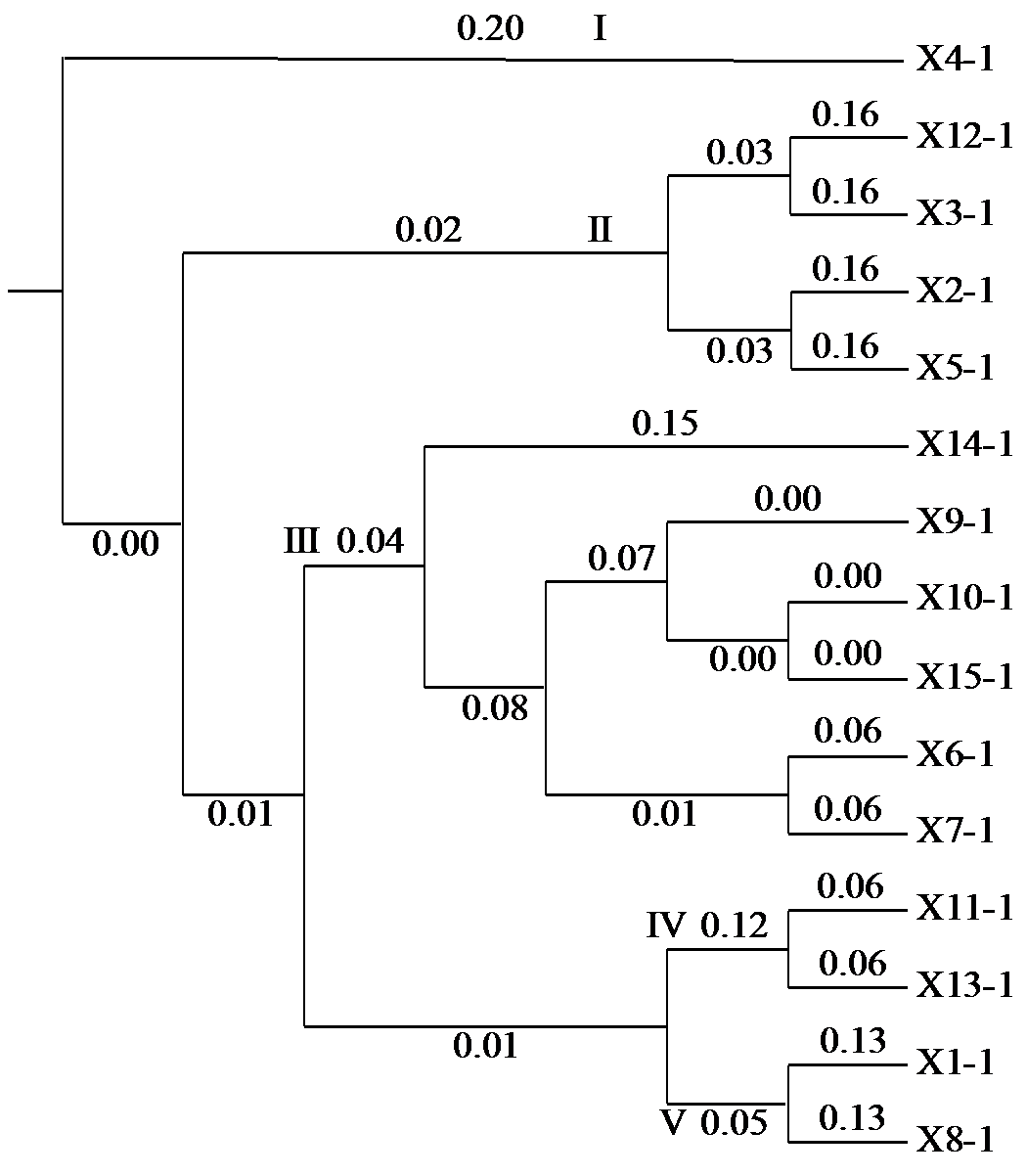
Genomic SSR Loci Mining and Genetic Diversity Analysis of Camellia oleifera Based on Genome Sequences
Dai Han1( ), Shen Tie1(
), Shen Tie1( ), Shi Taoxiong2, Li Ruiyuan1(
), Shi Taoxiong2, Li Ruiyuan1( )
)
- 1Guizhou Provincial Key Laboratory of Information and Computing Science, Guizhou Normal University, Guiyang 550001, Guizhou, China
2College of Life Sciences / Buckwheat Industry Technology Research Center, Guizhou Normal University, Guiyang 550001, Guizhou, China
| [1] | 赵欣, 宋家明, 张敬涛, 等. 基于SSR标记的海南油茶DNA指纹图谱构建. 分子植物育种,(2023-06-12) [2023-07-10].http://kns.cnki.net/kcms/detail/46.1068.S.20230612.1539.016.html. |
| [2] | 周文才, 温强, 杨军, 等. 油茶栽培品种SSR指纹图谱构建及聚类分析. 分子植物育种, 2017, 15(1):238-249. |
| [3] | 张恩慧, 王晓云, 覃子海, 等. 广西普通油茶种质资源遗传多样性的SSR分析. 广西植物, 2016, 36(7):806-811. |
| [4] | 李海波, 丁红梅, 陈友吾, 等. 12个油茶品种的SSR特征指纹鉴别. 中国粮油学报, 2017, 32(10):171-178. |
| [5] | 廖初琴. 赣南油茶EST-SSR分子标记的开发及遗传多样性分析. 广州: 广州大学, 2023. |
| [6] | 李海波, 王珊, 丁红梅, 等. 普通油茶转录组EST-SSR分子标记开发. 植物生理学报, 2017, 53(7):1267-1278. |
| [7] | 宋家明, 李欣窈, 张诗慧, 等. 基于转录组数据的海南油茶SSR分子标记的开发与评价. 分子植物育种, 2022, 20(20):6791- 6801. |
| [8] | 黎瑞源, 邢辉, 申铁. 四球茶转录组SSR位点信息分析. 安徽农业大学学报, 2017, 44(4):558-562. |
| [9] | 石桃雄, 黎瑞源, 潘凡, 等. 苦荞黄酮含量与SSR标记的关联分析. 福建农业学报, 2021, 36(8):884-891. |
| [10] | 蔡齐宗, 王佳蕊, 陈庆富, 等. 苦荞全基因组SSR位点鉴定及分子标记开发. 河南农业大学学报, 2022, 56(3):392-400. |
| [11] |
刘琳, 孙晓帅, 李乾, 等. 苹果SSR-PCR反应体系的建立与优化. 中国农学通报, 2021, 37(1):61-66.
doi: 10.11924/j.issn.1000-6850.casb20200100068 |
| [12] | 刘青青, 马玉花, 李雄杰, 等. 中国沙棘SSR-PCR反应体系的优化及引物开发. 基因组学与应用生物学, 2023, 42(10):1-15. |
| [13] | 靳雅惠. 桂郁金SSR-PCR反应体系的建立及优化. 陕西农业科学, 2020, 66(6):53-55. |
| [14] |
李振, 仲崇禄, 张勇, 等. 木麻黄SSR-PCR反应体系的建立与优化. 植物研究, 2021, 41(2):312-320.
doi: 10.7525/j.issn.1673-5102.2021.02.019 |
| [15] | He Z, Liu C, Wang X, et al. Assessment of genetic diversity in Camellia oleifera Abel. accessions using morphological traits and simple sequence repeat (SSR) markers. Breeding Science, 2020, 70(5):586-593. |
| [16] | Amarasinghe V, Carlson J E. The development of microsatellite DNA markers for genetic analysis in Douglas-fir. Canadian Journal of Forest Research, 2002, 32:1904-1915. |
| [17] | Wang P, Su J, Wu H, et al. Analysis of germplasm genetic diversity and construction of a core collection in Camellia oleifera C. Abel by integrating novel simple sequence repeat markers. Genetic Resources and Crop Evolution, 2023, 70(5):1517-1530. |
| [18] | Zhu Y Z, Liang D Y, Song Z J, et al. Genetic diversity analysis and core germplasm collection construction of Camellia oleifera based on fruit phenotype and SSR data. Genes, 2022, 13(12):2351. |
| [19] | 王德源, 杨冰, 朱亚艳, 等. 威宁短柱油茶转录组SSR位点信息分析. 分子植物育种,(2022-07-26) [2023-07-10].http://kns.cnki.net/kcms/detail/46.1068.S.20220726.1136.004.html. |
| [20] | 赵美容, 代惠萍, 倪珍珍, 等. 油茶种质遗传多样性研究进展. 园艺与种苗, 2016(3):31-32. |
| [21] | 田仟仟, 黄建建, 温强, 等. 油茶分子育种现状与趋势. 南方林业科学, 2021, 49(5):53-59. |
| [22] | He Z, Liu C, Wang X, et al. Assessment of genetic diversity in Camellia oleifera Abel. accessions using morphological traits and simple sequence repeat (SSR) markers: Research Papers. Breeding Science, 2020, 70(5):586-593. |
| [23] | Yan T, Gao L Z. Development and characterization of EST-SSR markers for Camellia reticulata. Applications in Plant Sciences, 2020, 8(5):e11348 |
| [24] | Alves A A, Pereira M V, Leão P A, et al. Advancing palm genomics by developing a high-density battery of molecular markers for Elaeis oleifera for future downstream applications,BMC Proceedings, 2014, 8(4):96. |
| [25] | 李琳琳, 黄万坚, 刘信凯, 等. 应用SSR分子标记技术鉴定张氏红山茶杂交F1代真实性的研究. 广东园林, 2014, 36(2):44-47. |
| [26] | 严和琴, 郑蔚, 代佳妮, 等. 基于SRAP分子标记的海南油茶品种遗传多样性分析. 分子植物育种, 2022, 20(6):1901-1908. |
| [27] | 安焱林, 李若冰, 王二赟, 等. 7个茶树全基因组SSR位点的鉴定与特征分析. 安徽农业大学学报, 2023, 50(1):58-63. |
| [28] | 林恩文, 林榕榕, 陈钦常, 等. 龙眼全基因组和转录本序列SSR位点的鉴定. 福建农林大学学报:自然科学版, 2022, 51(4):493-501. |
| [29] | 周文才, 温强, 杨军, 等. 油茶栽培品种SSR指纹图谱构建及聚类分析. 分子植物育种, 2017, 15(1):238-249. |
| [30] | 苗立祥, 杨肖芳, 张豫超, 等. 基于草莓全基因组SSR标记的开发和应用. 分子植物育种, 2021, 19(4):1210-1222. |
| [1] | Ma Hongzhen, Xu Haitao, Wang Yue, Feng Xiaoxi, Xu Bo, Zhang Jungang, Guo Haibin, Wang Youhua. Analysis of Genetic Diversity and Genetic Distance of Maize Inbred Lines Based on Phenotypic Traits of Husks [J]. Crops, 2024, 40(3): 54-63. |
| [2] | Quan Chengzhe, Li Shufang, Li Henan, Yu Wei, Jin Jinghua. Genetic Diversity Study of Phenotypic Traits of 73 Rice Varieties by Approved in Jinlin Province [J]. Crops, 2024, 40(3): 64-75. |
| [3] | Yang Enze, Xie Rui, Han Ping'an, Zhang Yonghu, Liu Jinchuan, Niu Suqing, Wen Rui, Wang Chunyong, Jin Xiaolei. Genetic Diversity and Comprehensive Evaluation of Phenotypic Traits of 162 Tartary Buckwheat Resources in Inner Mongolia [J]. Crops, 2024, 40(2): 15-22. |
| [4] | Ma Juan, Huang Lu, Yu Ting, Guo Guojun, Zhu Weihong, Liu Jingbao. Multi-Locus Genome-Wide Association Study and Genomic Prediction for General Combining Ability of Maize Ear Diameter [J]. Crops, 2024, 40(1): 31-39. |
| [5] | Liu Dan, Wang Jiayu, Feng Zhangli, Feng Bo, Chen Wenfu. Analysis on Genetic Diversity and Population Structure for Japonica Rice Varieties in Liaoning Province [J]. Crops, 2024, 40(1): 40-47. |
| [6] | Sun Yuantao, Long Wenjing, Li Yuan, Liu Tianpeng, Zhao Ganlin, Ding Guoxiang, Ni Xianlin. Genetic Diversity Analysis of 45 Glutinous Sorghum Germplasms Based on Major Agronomic Traits and SSR Markers [J]. Crops, 2024, 40(1): 57-64. |
| [7] | Wang Yueying, Fan Baojie, Cao Zhimin, Wang Yan, Su Qiuzhu, Zhang Zhixiao, Wang Shen, Shi Huiying, Shen Yingchao, Cheng Xuzhen, Liu Changyou, Tian Jing. Genetic Diversity Analysis of Landraces and Improved Varieties of Mung Bean by EST-SSR Markers [J]. Crops, 2024, 40(1): 73-79. |
| [8] | Qu Zhihua, Zhang Lili, Hu Yang, Qiao Haiming, Li Feng, Bai Wei. Agronomic Characteristics Evaluation on Introduced Flax Germplasm Resources [J]. Crops, 2023, 39(6): 47-53. |
| [9] | Zhao Feng, Bao Qijun, Pan Yongdong, Liu Xiaoning, Zhang Huayu, Niu Xiaoxia. Comprehensive Evaluation of Genetic Diversity in 70 Barley Germplasms [J]. Crops, 2023, 39(6): 54-61. |
| [10] | Yang Enze, Wang Shuyan, Liu Ruixiang, Shi Fengyuan, Zhang Jinhao, Li Jiana, Li Zhiwei, Guo Zhanbin. Genetic Diversity Analysis of Quinoa Germplasm Resources Based on SRAP [J]. Crops, 2023, 39(6): 79-85. |
| [11] | Zhang Shangpei, Yang Junxue, Luo Shiwu, Wang Yong, Zhang Xiaojuan, Cheng Bingwen. Genetic Diversity and Yielding Ability Analysis of Agronomic Traits in Broom Corn Millet [J]. Crops, 2023, 39(5): 37-42. |
| [12] | Gao Zhanning, Yang Yongqian, Wang Shujie, Feng Hui, Xue Zhenggang. Comprehensive Evaluation of 143 Barley Germplasm Resources [J]. Crops, 2023, 39(5): 59-65. |
| [13] | ChenZhikai , Hou Wanwei. Evaluation and Selection of Pisumsativum L. Germplasm Resources Based on Agronomic Traits [J]. Crops, 2023, 39(4): 38-43. |
| [14] | Li Qingfeng, Gao Jie, Peng Qiu. Genetic Diversity Analysis of Agronomic and Quality Characteristics of Amaranthus Resources in Guizhou Province [J]. Crops, 2023, 39(4): 60-64. |
| [15] | Li Hongsheng, Li Shaoxiang, Yang Zhonghui, Yang Jiali, Liu Kun, Xiong Shian, Li Fuqian, Guo Hui, Yang Mujun. Comparison ofPhenotype and Marker Detection in Seed Purity of Thermo-Photo Sensitive Two-Line WheatHybrids [J]. Crops, 2023, 39(4): 71-76. |
|
||