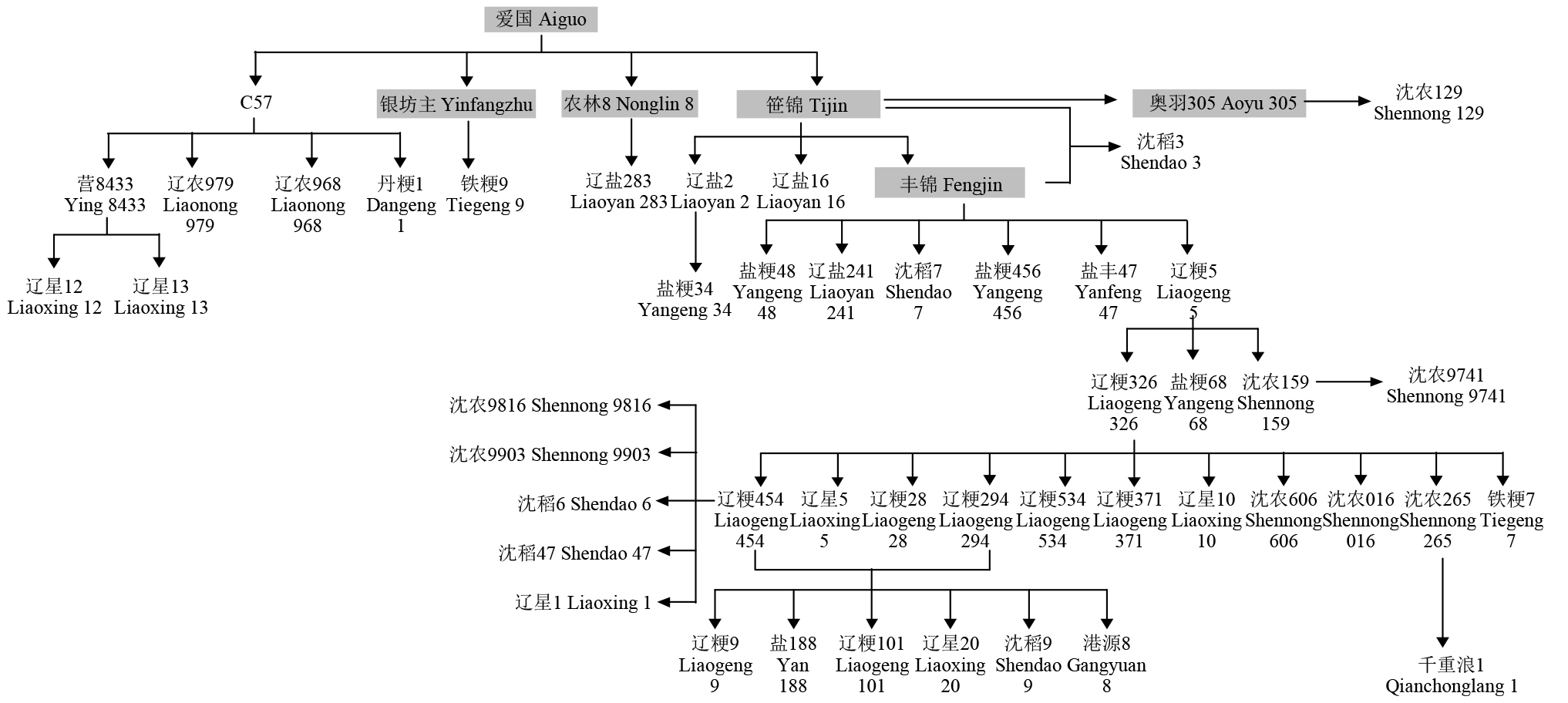
Crops ›› 2024, Vol. 40 ›› Issue (1): 40-47.doi: 10.16035/j.issn.1001-7283.2024.01.006
Previous Articles Next Articles
Analysis on Genetic Diversity and Population Structure for Japonica Rice Varieties in Liaoning Province
Liu Dan1( ), Wang Jiayu2(
), Wang Jiayu2( ), Feng Zhangli1, Feng Bo3, Chen Wenfu2
), Feng Zhangli1, Feng Bo3, Chen Wenfu2
- 1College of Biology and Agriculture of Zunyi Normal University, Zunyi 563006, Guizhou, China
2Rice Research Institute of Shenyang Agricultural University / Key Laboratory of Northeast Rice Biology and Genetic Breeding, Ministry of Agriculture and Rural Affairs, Shenyang 110866, Liaoning, China
3College of Resources and Environment of Zunyi Normal University, Zunyi 563006, Guizhou, China
| [1] | 国家水稻数据中心, 中国水稻品种及其系谱数据库. [2022-08- 10].http://www.ricedata.cn/variety/. |
| [2] |
Liu D, Wang J Y, Wang X X, et al. Genetic diversity and elite gene introgression reveal the japonica rice breeding in northern China. Journal of Integrative Agriculture, 2015, 14(5):811-822.
doi: 10.1016/S2095-3119(14)60898-4 |
| [3] |
李红宇, 侯昱铭, 陈英华, 等. 用SSR标记评估东北三省水稻推广品种的遗传多样性. 中国水稻科学, 2009, 23(4):383-390.
doi: 10.3969/j.issn.1001-7216.2009.04.08 |
| [4] | 张科, 魏海锋, 卓大龙, 等. 黑龙江省近年审定水稻品种基于SSR标记的遗传多样性分析. 植物遗传资源学报, 2016, 17(3):447-454. |
| [5] |
张佳琦, 郭宗珊, 刘长华, 等. 黑龙江省水稻品种的遗传多样性. 中国农学通报, 2022, 38(17):1-9.
doi: 10.11924/j.issn.1000-6850.casb2021-1183 |
| [6] | 孙健, 王敬国, 刘化龙, 等. 黑龙江省主栽水稻品种的遗传多样性分析. 作物杂志, 2011(1):63-66. |
| [7] | 张希瑞, 高文硕, 王敬国, 等. 吉林省水稻品种的遗传多样性及株型演化分析. 江苏农业学报, 2019, 35(3):497-505. |
| [8] |
Liu K, Muse S V. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics, 2005, 21(9):2128- 2129.
doi: 10.1093/bioinformatics/bti282 pmid: 15705655 |
| [9] |
Tamura K, Dudley J, Nei M, et al. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution, 2007, 24:1596-1599.
doi: 10.1093/molbev/msm092 pmid: 17488738 |
| [10] |
Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology, 2005, 14:2611-2620.
doi: 10.1111/j.1365-294X.2005.02553.x pmid: 15969739 |
| [11] |
张立娜, 曹桂兰, 韩龙植. 利用SSR标记揭示中国粳稻地方品种遗传多样性. 中国农业科学, 2012, 45(3):405-413.
doi: 10.3864/j.issn.0578-1752.2012.03.001 |
| [12] |
Salem K F M, Sallam A. Analysis of population structure and genetic diversity of Egyptian and exotic rice (Oryza sativa L.) genotypes. Comptes Rendus Biologies, 2016, 339(1):1-9.
doi: 10.1016/j.crvi.2015.11.003 |
| [13] |
Li W, Bian C M, Wei Y M, et al. Evaluation of genetic diversity of Sichuan common wheat landraces in China by SSR Markers. Journal of Integrative Agriculture, 2013, 12(9):1501-1511.
doi: 10.1016/S2095-3119(13)60418-9 |
| [14] |
Li L J, Yang K C, Pan G T, et al. Genetic diversity of maize populations developed by two kinds of recurrent selection methods investigated with SSR Markers. Agricultural Sciences in China, 2008, 7(9):1037-1045.
doi: 10.1016/S1671-2927(08)60144-3 |
| [15] | 李修平, 刘琪, 邵红, 等. 寒地水稻种质资源遗传多样性分析. 分子植物育种, 2021, 19(16):5528-5534. |
| [16] | 王敬国. 东北亚地区粳稻遗传多样性及主要株型性状与SSR标记的关联分析. 哈尔滨:东北林业大学, 2014. |
| [17] | 邵国军, 李玉福, 邱福林. 辽宁省水稻育种研究与进展. 辽宁农业科学, 1995(6):28-32. |
| [18] | 刘丹, 王嘉宇, 刘进, 等. 黑龙江省部分育成粳稻资源的遗传多样性分析. 植物遗传资源学报, 2019, 20(1):60-68. |
| [19] | 程芳艳, 李春光, 刘永巍, 等. 寒地部分粳稻的遗传多样性及遗传结构分析. 沈阳农业大学学报, 2014, 45(6):649-654. |
| [1] | Sun Yuantao, Long Wenjing, Li Yuan, Liu Tianpeng, Zhao Ganlin, Ding Guoxiang, Ni Xianlin. Genetic Diversity Analysis of 45 Glutinous Sorghum Germplasms Based on Major Agronomic Traits and SSR Markers [J]. Crops, 2024, 40(1): 57-64. |
| [2] | Wang Yueying, Fan Baojie, Cao Zhimin, Wang Yan, Su Qiuzhu, Zhang Zhixiao, Wang Shen, Shi Huiying, Shen Yingchao, Cheng Xuzhen, Liu Changyou, Tian Jing. Genetic Diversity Analysis of Landraces and Improved Varieties of Mung Bean by EST-SSR Markers [J]. Crops, 2024, 40(1): 73-79. |
| [3] | Zhao Feng, Bao Qijun, Pan Yongdong, Liu Xiaoning, Zhang Huayu, Niu Xiaoxia. Comprehensive Evaluation of Genetic Diversity in 70 Barley Germplasms [J]. Crops, 2023, 39(6): 54-61. |
| [4] | Yang Enze, Wang Shuyan, Liu Ruixiang, Shi Fengyuan, Zhang Jinhao, Li Jiana, Li Zhiwei, Guo Zhanbin. Genetic Diversity Analysis of Quinoa Germplasm Resources Based on SRAP [J]. Crops, 2023, 39(6): 79-85. |
| [5] | Liu Qiuyuan, Li Meng, Gao Yangguang, Shi Mengyu, Wei Yunfei, Ji Xin, Li Li, Liu Yali, Wang Fujuan. Effects of Different Nitrogen Fertilization Patterns on Yield and Quality of Conventional Japonica Rice under Reduced Nitrogen [J]. Crops, 2023, 39(5): 131-137. |
| [6] | Zhang Shangpei, Yang Junxue, Luo Shiwu, Wang Yong, Zhang Xiaojuan, Cheng Bingwen. Genetic Diversity and Yielding Ability Analysis of Agronomic Traits in Broom Corn Millet [J]. Crops, 2023, 39(5): 37-42. |
| [7] | Gao Zhanning, Yang Yongqian, Wang Shujie, Feng Hui, Xue Zhenggang. Comprehensive Evaluation of 143 Barley Germplasm Resources [J]. Crops, 2023, 39(5): 59-65. |
| [8] | Zhai Jing, Yang Shengming, Wang Yuzhen, Shi Linlin. Effects of Multi-Year Organic Fertilizer Application on Starch Physicochemical Properties of Mid-Mature Soft Japonica Rice [J]. Crops, 2023, 39(5): 91-97. |
| [9] | ChenZhikai , Hou Wanwei. Evaluation and Selection of Pisumsativum L. Germplasm Resources Based on Agronomic Traits [J]. Crops, 2023, 39(4): 38-43. |
| [10] | Li Qingfeng, Gao Jie, Peng Qiu. Genetic Diversity Analysis of Agronomic and Quality Characteristics of Amaranthus Resources in Guizhou Province [J]. Crops, 2023, 39(4): 60-64. |
| [11] | Li Hongsheng, Li Shaoxiang, Yang Zhonghui, Yang Jiali, Liu Kun, Xiong Shian, Li Fuqian, Guo Hui, Yang Mujun. Comparison ofPhenotype and Marker Detection in Seed Purity of Thermo-Photo Sensitive Two-Line WheatHybrids [J]. Crops, 2023, 39(4): 71-76. |
| [12] | Chen Cuiping, Yan Dianhai, Zhang Shumiao, Zuo Haonan, Gao Sen, Liu Yang. Fingerprint Construction and Genetic Diversity Analysis of Quinoa Based on SSR Markers [J]. Crops, 2023, 39(3): 35-42. |
| [13] | Song Yun, Zhang Xinrui, He Jiaxin, Li Zheng, Sun Zhe, Li Aoxuan, Qiao Yonggang. Genetic Diversity Analysis of Sophora flavescens Ait. Germplasm Resources Based on cpSSR Markers [J]. Crops, 2023, 39(1): 30-37. |
| [14] | Guo Huanle, Tang Bin, Li Han, Cao Zhongyang, Zeng Qiang, Liu Liangwu, Chen Zhihui. Comprehensive Evaluation of Phenotypic Traits and Classification of Maize Landraces in Hunan Province [J]. Crops, 2022, 38(6): 33-41. |
| [15] | Zhao Xiaoqin, Jia Ruiling, Liu Junxiu, Liu Yanming, Wen Yinhua, Shi Lili, Zhang Juanning, Ma Ning. Agronomic Traits and Genetic Diversity Analysis of 120 Foxtail Millet Germplasms [J]. Crops, 2022, 38(6): 61-69. |
|
||