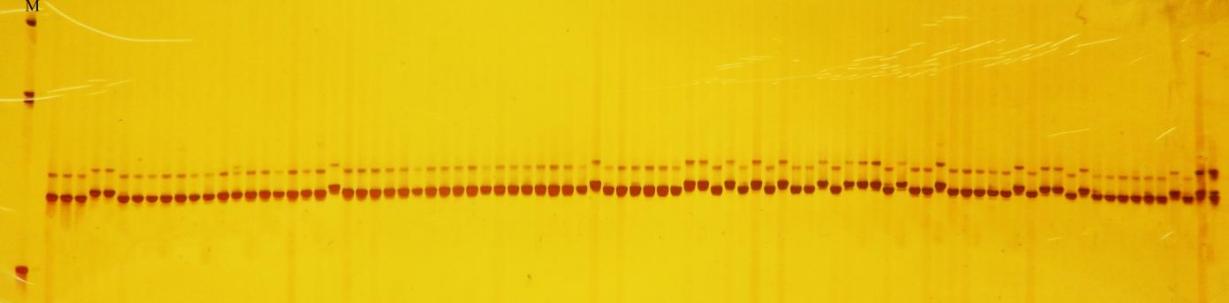
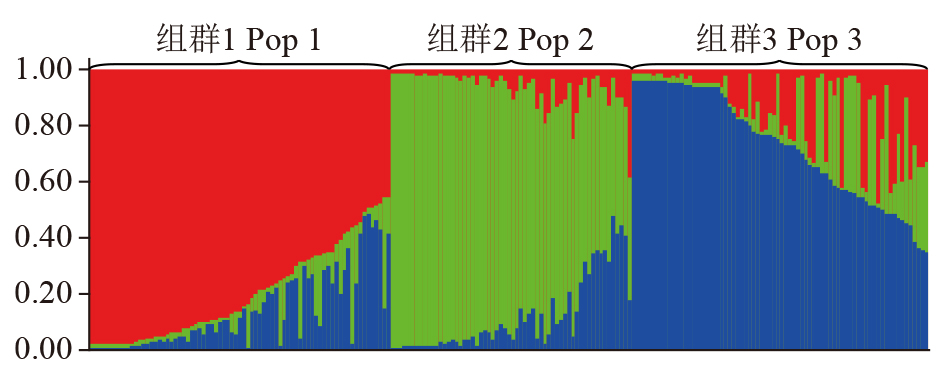
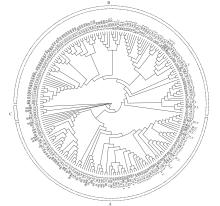
Crops ›› 2024, Vol. 40 ›› Issue (1): 73-79.doi: 10.16035/j.issn.1001-7283.2024.01.010
Previous Articles Next Articles
Genetic Diversity Analysis of Landraces and Improved Varieties of Mung Bean by EST-SSR Markers
Wang Yueying1,2( ), Fan Baojie1, Cao Zhimin1, Wang Yan1, Su Qiuzhu1, Zhang Zhixiao1, Wang Shen1, Shi Huiying1, Shen Yingchao1, Cheng Xuzhen3, Liu Changyou1(
), Fan Baojie1, Cao Zhimin1, Wang Yan1, Su Qiuzhu1, Zhang Zhixiao1, Wang Shen1, Shi Huiying1, Shen Yingchao1, Cheng Xuzhen3, Liu Changyou1( ), Tian Jing1(
), Tian Jing1( )
)
- 1Institute of Food and Oil Crops, Hebei Academy of Agricultural and Forestry Sciences / Hebei Laboratory of Crop Genetic Breeding, Shijiazhuang 050031, Hebei, China
2China National Rice Research Institute, Chinese Academy of Agricultural Sciences / State Key Laboratory of Rice Biology, Hangzhou 310006
3Zhejiang, China
3Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100081, China
| [1] |
Chen H L, Qiao L, Wang L X, et al. Assessment of genetic diversity and population structure of mung bean (Vigna radiata) germplasm using EST-based and genomic SSR markers. Gene, 2015, 566(2):175-183.
doi: 10.1016/j.gene.2015.04.043 pmid: 25895480 |
| [2] |
Chen H L, Wang L X, Wang S H, et al. Transcriptome sequencing of mung bean (Vigna radiate L.) genes and the identification of EST-SSR markers. PLoS ONE, 2015, 10(4):e0120273.
doi: 10.1371/journal.pone.0120273 |
| [3] |
Kim S K, Nair R M, Lee J, et al. Genomic resources in mungbean for future breeding programs. Frontiers in Plant Science, 2015, 6:626.
doi: 10.3389/fpls.2015.00626 pmid: 26322067 |
| [4] | 程须珍. 绿豆种质资源挖掘与新品种创制应用研究取得重要进展. 植物遗传资源学报, 2015, 16(4):674. |
| [5] | 刘慧. 我国绿豆生产现状和发展前景. 农业生产展望, 2012, 8(6):36-39. |
| [6] | 刘浩, 周闲容, 于晓娜, 等. 作物种质资源品质性状鉴定评价现状与展望. 植物遗传资源学报, 2014, 15(1):215-221. |
| [7] | 任红晓, 姜翠棉, 高运青, 等. 中国传统名优绿豆种质资源表型性状形态多样性. 作物杂志, 2017(1):44-47. |
| [8] |
张盼盼, 张洪鹏, 张敬禹, 等. 绿豆种质资源农艺性状的因子分析及综合评价. 中国农学通报, 2017, 33(6):34-41.
doi: 10.11924/j.issn.1000-6850.casb16040131 |
| [9] |
乔玲, 陈红霖, 王丽侠, 等. 国外绿豆种质资源农艺性状的遗传多样性分析. 植物遗传资源学报, 2015, 16(5):986-993.
doi: 10.13430/j.cnki.jpgr.2015.05.009 |
| [10] | 朱慧珺. 山西省绿豆种质资源表型性状遗传多样性分析. 晋中:山西农业大学, 2015. |
| [11] | 张毅华, 张耀文, 张泽燕. 绿豆种质资源表型性状多样性分析. 农学学报, 2013, 3(1):15-19. |
| [12] | 王建波. ISSR分子标记及其在植物遗传学研究中的应用. 遗传, 2002, 24(5):613-616. |
| [13] |
Chen H L, Liu L P, Wang L X, et al. Development of SSR markers and assessment of genetic diversity of adzuki bean in the Chinese germplasm collection. Molecular Breeding, 2017, 37(5):66.
doi: 10.1007/s11032-017-0662-4 |
| [14] |
Sangiri C, Kaga A, Tomooka N, et al. Genetic diversity of the mungbean (Vigna radiata, Leguminosae) genepool on the basis of microsatellite analysis. Australian Journal of Botany, 2008, 55 (8):837-847.
doi: 10.1071/BT07105 |
| [15] |
Wang L X, Elbaidouri M, Abernathy B, et al. Distribution and analysis of SSR in mung bean (Vigna radiata L.) genome based on an SSR-enriched library. Molecular Breeding, 2015, 35(1):25.
doi: 10.1007/s11032-015-0259-8 |
| [16] |
刘岩, 程须珍, 王丽侠, 等. 基于SSR标记的中国绿豆种质资源遗传多样性研究. 中国农业科学, 2013, 46(20):4197-4209.
doi: 10.3864/j.issn.0578-1752.2013.20.003 |
| [17] |
赵雪英, 王宏民, 李赫, 张耀文. 绿豆种质资源的ISSR遗传多样性分析. 植物遗传资源学报, 2015, 16(6):1277-1282.
doi: 10.13430/j.cnki.jpgr.2015.06.020 |
| [18] | 王丽侠, 程须珍, 王素华, 等. 小豆SSR引物在绿豆基因组中的通用性分析. 作物学报, 2009, 35(5):816-820. |
| [19] |
任红晓, 程须珍, 徐东旭, 等. 应用SSR标记分析中国北方名优绿豆的遗传多样性. 植物遗传资源学报, 2015, 16(2):395-399.
doi: 10.13430/j.cnki.jpgr.2015.02.028 |
| [20] | 杨新泉, 刘鹏, 韩宗福, 等. 普通小麦Genomic-SSR和EST-SSR分子标记遗传差异及其与系谱遗传距离的比较研究. 遗传学报, 2005, 32(4):406-416. |
| [21] |
Porebski S, Bailey L G, Baum B R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Molecular Biology Reporter, 1997, 15(1):8-15.
doi: 10.1007/BF02772108 |
| [22] |
Liu C Y, Fan B J, Cao Z M, et al. A deep sequencing analysis of transcriptomes and the development of EST-SSR markers in mungbean (Vigna radiata). Journal of Genetics, 2016, 95(3):527.
pmid: 27659323 |
| [23] |
Evanno G S, Regnaut S J, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Molecular Ecology, 2005, 14(8):2611-2620.
doi: 10.1111/j.1365-294X.2005.02553.x pmid: 15969739 |
| [24] | 姜伟, 何觉民, 陆建农, 等. 中国主要棉花栽培群体的遗传多样性分析. 农业生物技术学报, 2008, 16(1):127-133. |
| [25] | 宫慧慧, 谢华, 马荣才, 等. 利用SSR分析小豆种质遗传多样性. 农业生物技术学报, 2008, 16(5):872-880. |
| [26] |
王丽侠, 程须珍, 王素华, 等. 应用SSR标记对小豆种质资源的遗传多样性分析. 作物学报, 2009, 35(10):1858-1865.
doi: 10.3724/SP.J.1006.2009.01858 |
| [27] |
徐宁, 程须珍, 王丽侠, 等. 用于中国小豆种质资源遗传多样性分析SSR分子标记筛选及应用. 作物学报, 2009, 35(2):219-227.
doi: 10.3724/SP.J.1006.2009.00219 |
| [28] |
Cho Y G, Ishii T, Temnykh S, et al. Diversity of microsatellites derived from genomic libraries and GenBank sequences in rice (Oryza sativa L.). Theoretical and Applied Genetics, 2000, 100 (5):713-722.
doi: 10.1007/s001220051343 |
| [1] | Liu Dan, Wang Jiayu, Feng Zhangli, Feng Bo, Chen Wenfu. Analysis on Genetic Diversity and Population Structure for Japonica Rice Varieties in Liaoning Province [J]. Crops, 2024, 40(1): 40-47. |
| [2] | Sun Yuantao, Long Wenjing, Li Yuan, Liu Tianpeng, Zhao Ganlin, Ding Guoxiang, Ni Xianlin. Genetic Diversity Analysis of 45 Glutinous Sorghum Germplasms Based on Major Agronomic Traits and SSR Markers [J]. Crops, 2024, 40(1): 57-64. |
| [3] | Qu Zhihua, Zhang Lili, Hu Yang, Qiao Haiming, Li Feng, Bai Wei. Agronomic Characteristics Evaluation on Introduced Flax Germplasm Resources [J]. Crops, 2023, 39(6): 47-53. |
| [4] | Zhao Feng, Bao Qijun, Pan Yongdong, Liu Xiaoning, Zhang Huayu, Niu Xiaoxia. Comprehensive Evaluation of Genetic Diversity in 70 Barley Germplasms [J]. Crops, 2023, 39(6): 54-61. |
| [5] | Yang Enze, Wang Shuyan, Liu Ruixiang, Shi Fengyuan, Zhang Jinhao, Li Jiana, Li Zhiwei, Guo Zhanbin. Genetic Diversity Analysis of Quinoa Germplasm Resources Based on SRAP [J]. Crops, 2023, 39(6): 79-85. |
| [6] | Zhang Shangpei, Yang Junxue, Luo Shiwu, Wang Yong, Zhang Xiaojuan, Cheng Bingwen. Genetic Diversity and Yielding Ability Analysis of Agronomic Traits in Broom Corn Millet [J]. Crops, 2023, 39(5): 37-42. |
| [7] | Gao Zhanning, Yang Yongqian, Wang Shujie, Feng Hui, Xue Zhenggang. Comprehensive Evaluation of 143 Barley Germplasm Resources [J]. Crops, 2023, 39(5): 59-65. |
| [8] | ChenZhikai , Hou Wanwei. Evaluation and Selection of Pisumsativum L. Germplasm Resources Based on Agronomic Traits [J]. Crops, 2023, 39(4): 38-43. |
| [9] | Li Qingfeng, Gao Jie, Peng Qiu. Genetic Diversity Analysis of Agronomic and Quality Characteristics of Amaranthus Resources in Guizhou Province [J]. Crops, 2023, 39(4): 60-64. |
| [10] | Chen Cuiping, Yan Dianhai, Zhang Shumiao, Zuo Haonan, Gao Sen, Liu Yang. Fingerprint Construction and Genetic Diversity Analysis of Quinoa Based on SSR Markers [J]. Crops, 2023, 39(3): 35-42. |
| [11] | Song Yun, Zhang Xinrui, He Jiaxin, Li Zheng, Sun Zhe, Li Aoxuan, Qiao Yonggang. Genetic Diversity Analysis of Sophora flavescens Ait. Germplasm Resources Based on cpSSR Markers [J]. Crops, 2023, 39(1): 30-37. |
| [12] | Huang Guibin, Guan Yaobing, Niu Yongqi, Zhou Lilei, Zhao Yongfeng. Comprehensive Evaluation of 12 Major Agronomic Traits of 103 Chickpea Germplasm Resources [J]. Crops, 2023, 39(1): 6-13. |
| [13] | Hou Xue, Chen Yujie, Li Chunmiao, Fang Shumei, Liang Xilong, Zheng Dianfeng. Regulating Effects of Prohexadione-Calcium on the Growth of Mung Bean Seedlings under Saline-Alkali Stress [J]. Crops, 2022, 38(6): 174-180. |
| [14] | Guo Huanle, Tang Bin, Li Han, Cao Zhongyang, Zeng Qiang, Liu Liangwu, Chen Zhihui. Comprehensive Evaluation of Phenotypic Traits and Classification of Maize Landraces in Hunan Province [J]. Crops, 2022, 38(6): 33-41. |
| [15] | Li Wangsheng, Wang Xueqian, Yin Xilong, Shi Yang, Liu Dali, Tan Wenbo, Xing Wang. Comprehensive Evaluation of Drought Tolerance of Sugar Beet Germplasms at Seedling Stage [J]. Crops, 2022, 38(6): 54-60. |
|
||