Crops ›› 2025, Vol. 41 ›› Issue (2): 66-73.doi: 10.16035/j.issn.1001-7283.2025.02.009
Previous Articles Next Articles
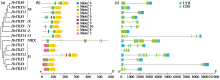
Identification and Bioinformatics Analysis of the Typical Thioredoxin (TRX) Gene Family in Barley
Chen Hu( ), Gao Yuan, Sun Jiameng, Yu Peng, Xiao Hongwu, Zhang Haitao(
), Gao Yuan, Sun Jiameng, Yu Peng, Xiao Hongwu, Zhang Haitao( )
)
- College of Agronomy, Anhui Agricultural University, Hefei 230036, Anhui, China
| [1] |
Haas M, Schreiber M, Mascher M. Domestication and crop evolution of wheat and barley: Genes, genomics, and future directions. Journal of Integrative Plant Biology, 2019, 61(3):204-225.
doi: 10.1111/jipb.12737 |
| [2] | Geng L, Li M D, Zhang G D, et al. Barley: a potential cereal for producing healthy and functional foods. Food Quality and Safety, 2022, 6(2):142-154. |
| [3] | Sakellariou M, Mylona P V. New uses for traditional crops: the case of barley biofortification. Agronomy, 2020, 10(12):1964. |
| [4] | Lemaire S D, Miginiac-Maslow M. The thioredoxin superfamily in Chlamydomonas reinhardtii. Photosynthesis Research, 2004,82:203-220. |
| [5] |
Arnér E S J, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. European Journal of Biochemistry, 2000, 267(20):6102-6109.
doi: 10.1046/j.1432-1327.2000.01701.x pmid: 11012661 |
| [6] |
Liu J, Liu B, Feng D R, et al. Evidence for a role of chloroplastic m-type thioredoxins in the biogenesis of photosystem II in Arabidopsis. Plant Physiology, 2013, 163(4):1710-1728.
doi: 10.1104/pp.113.228353 pmid: 24151299 |
| [7] | Zhou J F, Song T Q, Zhou H W, et al. Genome-wide identification, characterization, evolution, and expression pattern analyses of the typical thioredoxin gene family in wheat (Triticum aestivum L.). Frontiers in Plant Science, 2022,13:1020584. |
| [8] | Jacquot J P, Vidal J, Gadal P, et al. Evidence for the existence of several enzyme-specific thioredoxins in plants. FEBS Letters, 1978, 96(2):243-246. |
| [9] | Wolosiuk R A, Buchanan B B. Thioredoxin and glutathione regulate photosynthesis in chloroplasts. Nature, 1977, 266(5602):565-567. |
| [10] |
Okegawa Y, Motohashi K. Chloroplastic thioredoxin m functions as a major regulator of Calvin cycle enzymes during photosynthesis in vivo. The Plant Journal, 2015, 84(5):900-913.
doi: 10.1111/tpj.13049 pmid: 26468055 |
| [11] |
Barajas-López J D, Serrato A J, Cazalis R, et al. Circadian regulation of chloroplastic f and m thioredoxins through control of the CCA 1 transcription factor. Journal of Experimental Botany, 2011, 62(6):2039-2051.
doi: 10.1093/jxb/erq394 pmid: 21196476 |
| [12] | Buchanan B B. The path to thioredoxin and redox regulation in chloroplasts. Annual Review of Plant Biology, 2016,67:1-24. |
| [13] | Gelhaye E, Rouhier N, Gérard J, et al. A specific form of thioredoxin h occurs in plant mitochondria and regulates the alternative oxidase. Proceedings of the National Academy of Sciences of the United States of America, 2004, 101(40):14545-14550. |
| [14] | Laloi C, Rayapuram N, Chartier Y, et al. Identification and characterization of a mitochondrial thioredoxin system in plants. Proceedings of the National Academy of Sciences of the United States of America, 2001, 98(24):14144-14149. |
| [15] | Collin V, Issakidis-Bourguet E, Marchand C, et al. The Arabidopsis plastidial thioredoxins: new functions and new insights into specificity. Journal of Biological Chemistry, 2003, 278(26):23747-23752. |
| [16] | Collin V, Lamkemeyer P, Miginiac-Maslow M, et al. Characterization of plastidial thioredoxins from Arabidopsis belonging to the new y-type. Plant Physiology, 2004, 136(4):4088-4095. |
| [17] | Arsova B, Hoja U, Wimmelbacher M, et al. Plastidial thioredoxin z interacts with two fructokinase-like proteins in a thiol- dependent manner: evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana. The Plant Cell, 2010, 22(5):1498-1515. |
| [18] |
Geigenberger P, Thormählen I, Daloso D M, et al. The unprecedented versatility of the plant thioredoxin system. Trends in Plant Science, 2017, 22(3):249-262.
doi: S1360-1385(16)30221-7 pmid: 28139457 |
| [19] |
Nikkanen L, Rintamäki E. Chloroplast thioredoxin systems dynamically regulate photosynthesis in plants. Biochemical Journal, 2019, 476(7):1159-1172.
doi: 10.1042/BCJ20180707 pmid: 30988137 |
| [20] |
Laughner B J, Sehnke P C, Ferl R J. A novel nuclear member of the thioredoxin superfamily. Plant Physiology, 1998, 118(3):987-996.
pmid: 9808743 |
| [21] |
Funato Y, Hayashi T, Irino Y, et al. Nucleoredoxin regulates glucose metabolism via phosphofructokinase 1. Biochemical and Biophysical Research Communications, 2013, 440(4):737-742.
doi: 10.1016/j.bbrc.2013.09.138 pmid: 24120946 |
| [22] | Chen C J, Chen H, Zhang Y, et al. TBtools:an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 2020, 13(8):1194-1202. |
| [23] |
Sun L J, Ren H Y, Liu R X, et al. An h-type thioredoxin functions in tobacco defense responses to two species of viruses and an abiotic oxidative stress. Molecular Plant-Microbe Interactions, 2010, 23(11):1470-1485.
doi: 10.1094/MPMI-01-10-0029 pmid: 20923353 |
| [24] |
Havelda Z, Várallyay É, Válóczi A, et al. Plant virus infection‐induced persistent host gene downregulation in systemically infected leaves. The Plant Journal, 2008, 55(2):278-288.
doi: 10.1111/j.1365-313X.2008.03501.x pmid: 18397378 |
| [25] | Valerio C, Costa A, Marri L, et al.Thioredoxin-regulated β-amylase (BAM1) triggers diurnal starch degradation in guard cells, and in mesophyll cells under osmotic stress. Journal of Experimental Botany, 2011, 62(2):545-555. |
| [26] | Moon H, Lee B, Choi G, et al. NDP kinase 2 interacts with two oxidative stress-activated MAPKs to regulate cellular redox state and enhances multiple stress tolerance in transgenic plants. Proceedings of the National Academy of Sciences of the United States of America, 2003, 100(1):358-363. |
| [27] |
Xie G, Kato H, Sasaki K, et al. A cold-induced thioredoxin h of rice, OsTrx23, negatively regulates kinase activities of OsMPK3 and OsMPK6 in vitro. FEBS Letters, 2009, 583(17):2734-2738.
doi: 10.1016/j.febslet.2009.07.057 pmid: 19665023 |
| [28] | Zhang S X, Yu Y, Song T Q, et al. Genome-wide identification of foxtail millet’s TRX family and a functional analysis of SiNRX1 in response to drought and salt stresses in transgenic Arabidopsis. Frontiers in Plant Science, 2022,13:946037. |
| [29] |
Nuruzzaman M, Sharoni A M, Satoh K, et al. The thioredoxin gene family in rice: Genome-wide identification and expression profiling under different biotic and abiotic treatments. Biochemical and Biophysical Research Communications, 2012, 423(2):417-423.
doi: 10.1016/j.bbrc.2012.05.142 pmid: 22683629 |
| [30] | Elasad M, Wei H L, Wang H T, et al. Genome-wide analysis and characterization of the TRX gene family in upland cotton. Tropical Plant Biology, 2018, 11(3/4):119-130. |
| [31] | Zhang J R, Zhao T, Yan F D, et al. Genome-wide identification and expression analysis of Thioredoxin (Trx) genes in seed development of vitis vinifera. Journal of Plant Growth Regulation, 2022, 41(7):3030-3045. |
| [32] | Li X, Su G J, Ntambiyukuri A, et al. Genome-wide identification and expression analysis of the AhTrx family genes in peanut. Biologia Plantarum, 2022, 66(1):112-122. |
| [33] | Bhurta R, Hurali D T, Tyagi S, et al. Genome-wide identification and expression analysis of the thioredoxin (trx) gene family reveals its role in leaf rust resistance in wheat (Triticum aestivum L.). Frontiers in Genetics, 2022,13:563. |
| [34] | Hägglund P, Björnberg O, Navrot N, et al. The barley grain thioredoxin system-an update. Frontiers in Plant Science, 2013,4:151. |
| [1] | Zhang Jun, Cai Suyun, Xu Zihao, Hou Lei, He Runli, Yin Guifang, Wang Lihua, Wang Yanqing, Lu Wenjie, Sun Daowang. Cloning, Bioinformatics and Expression Analysis of FtERF Gene in Fagopyrum tataricum [J]. Crops, 2024, 40(2): 23-29. |
| [2] | Zhang Qian, Ren Wen, Zhao Bingbing, Zhou Miaoyi, Li Hanshuai, Liu Ya, Du Hewei. Cloning and Bioinformatics Analysis of ZmMAPKKK21 Gene in Maize [J]. Crops, 2024, 40(2): 30-39. |
| [3] | Zhang Yu, Yang Wenjing, Liu Xuan, Nie Fengjie, Zhang Li, Shi Lei, Zhang Guohui, Guo Zhiqian, Gong Lei. Cloning and Expression Analysis of Potato StCWIN1 Gene Promoter and Its Role under Drought Stress [J]. Crops, 2024, 40(2): 54-61. |
| [4] | Lü Baolian, Yang Yuxin, Cui Licao, Shi Feng, Ma Liang, Kong Xiuying, Zhang Lichao, Ni Zhiyong. Identification of bHLH Family Transcription Factors of Wheat and Expression Analysis under Salt Stress [J]. Crops, 2024, 40(1): 65-72. |
| [5] | Wu Ying, Hu Die, Li Ting, Duan Qianyuan, Wei Ningning, Zhang Xinghua, Xu Shutu, Xue Jiquan. Analysis of WRKY Transcription Factor IIc Subfamily in Maize and Its Expression Profile under Drought [J]. Crops, 2024, 40(1): 80-89. |
| [6] | Zhao Pengpeng, Li Luhua, Ren Mingjian, An Chang, Hong Dingli, Li Xin, Xu Ruhong. Bioinformatics and Expression Analysis of GzCIPK7-5B Gene in Wheat [J]. Crops, 2023, 39(4): 77-84. |
| [7] | Qiu Kaihua, Fang Shumei, Liang Xilong. Functional Analysis of SRRM1-Like Transcription Factor of Magnaporthe grisea [J]. Crops, 2023, 39(3): 246-253. |
| [8] | Meng Yaxuan, Yao Xuhang, Sun Yingqi, Zhao Xinyue, Wang Fengxia, Weng Qiaoyun, Liu Yinghui. Identification and Bioinformatics Analysis of DGAT Gene Family in Cereal Crops [J]. Crops, 2023, 39(1): 20-29. |
| [9] | Zhou Fei. Bioinformatics and Expression Analysis of HaLACS7 Gene in Sunflower [J]. Crops, 2022, 38(3): 104-108. |
| [10] | Yang Xiaolin, Duan Ying, Cai Suyun, He Runli, Yin Guifang, Wang Yanqing, Lu Wenjie, Sun Daowang, Wang Lihua. Molecular Cloning and Bioinformatics Analyzing of Laccase in Fagopyrum tataricum [J]. Crops, 2022, 38(3): 73-79. |
| [11] | Lu Ping, Kang Qingfang, Zhao Mengyao, Zhang Fengjie, Wu Qiangqiang, Ma Fangfang, Wang Yushen, Han Yuanhuai, Wang Xingchun, Li Xueyin. Identification and Functional Analysis of miR169 Family and Its Target Genes in Setaria italica [J]. Crops, 2022, 38(2): 54-63. |
| [12] | Yin Guifang, Duan Ying, Yang Xiaolin, Cai Suyun, Wang Yanqing, Lu Wenjie, Sun Daowang, He Runli, Wang Lihua. Cloning and Bioinformatics Analysis of FtC4H Gene from Tartary Buckwheat [J]. Crops, 2022, 38(1): 77-83. |
| [13] | Wang Zhilong, Xue Yinghong, Hao Yueru, Liu Baoling, Yuan Lixia, Xue Jin'ai, Li Runzhi. Identification and Expression Analysis of Oil-Related Transcription actor CsLEC2 Gene Family in Camelina sativa (L.) Crantz [J]. Crops, 2020, 36(5): 23-32. |
| [14] | Xu Yuanyuan, Zhao Peng, Hong Quanchun, Zhu Xiaoqin, Pei Dongli. Isolation and Expression Analysis of Transcription Factor Gene TaMYB70 in Wheat [J]. Crops, 2020, 36(4): 84-90. |
| [15] | Zhao Xunchao,Xu Jingyu,Gai Shengnan,Wei Yulei,Xu Xiaoxuan,Ding Dong,Liu Meng,Zhang Jinjie,Shao Wenjing. Identification of Stearyl -ACP Desaturase Gene (SbSAD) Family and Their Expression Analysis at Different Developmental Stages in Sorghum [J]. Crops, 2020, 36(2): 20-27. |
|