Crops ›› 2018, Vol. 34 ›› Issue (6): 53-57.doi: 10.16035/j.issn.1001-7283.2018.06.009
Previous Articles Next Articles
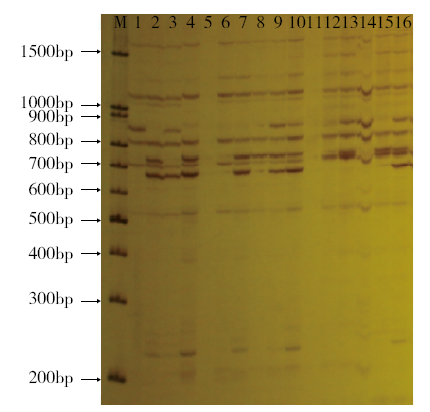
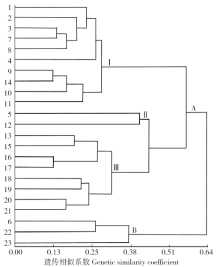
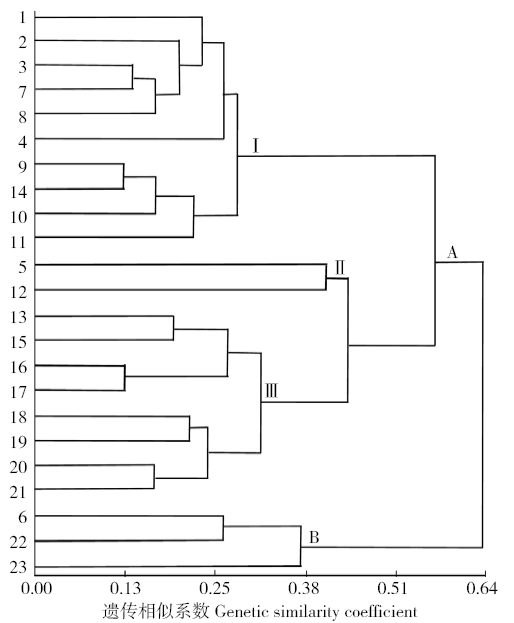
Genetic Diversity of Flax Germplasm Resources in Inner Mongolia
Yi Liuxi1,Siqin Bateer1,Gao Fengyun1,Zhou Yu1,Jia Xiaoyun1,Chen Mingzhe2,Zhang Hui1,Jia Haibin3
- 1 Biotechnology research center of the Inner Mongolia Academy of Agriculture and Husbandry Sciences, Huhhot 010031, Inner Mongolia, China
2 Xilinguole Meng Institute of Agricultural Sciences, Xilinhaote 026000, Inner Mongolia, China
3 Wulanchabu Academy of Agriculture and Animal Husbandry, Jining 012000, Inner Mongolia, China
| [1] |
伊六喜, 斯钦巴特尔, 贾霄云 ,等. 胡麻种质资源、育种及遗传研究进展. 中国麻业科学, 2017,39(2):81-87.
doi: 10.3969/j.issn.1671-3532.2017.02.006 |
| [2] | 伊六喜, 斯钦巴特尔, 张辉 ,等. 胡麻种质资源遗传多样性及亲缘关系的SRAP分析. 西北植物学报, 2017,37(10):1941-1950. |
| [3] | 伊六喜, 萨如拉, 张辉 , 等. 胡麻种子产量与主要农艺性状的多重分析. 安徽农业科学, 2018,46(6):33-36. |
| [4] | 李明, 姜硕, 郑东泽 , 等. 亚麻SRAP标记连锁图谱的构建及3个数量性状的定位. 东北农业大学学报, 2014,45(2):12-18. |
| [5] |
安泽山, 严兴初, 党占海 , 等. 利用SRAP标记分析胡麻资源遗传多样性. 西南农业学报, 2014,27(2):530-534.
doi: 10.3969/j.issn.1001-4829.2014.02.013 |
| [6] | 伊六喜 , 斯钦巴特尔,张辉,等. 胡麻核心种质资源表型变异及SRAP分析. 中国油料作物学报, 2017,39(6):794-804. |
| [7] |
郝荣楷, 严兴初, 党占海 , 等. 我国胡麻育成品种的遗传多样性分析. 中国油料作物学报, 2014,36(3):334-342.
doi: 10.7505/j.issn.1007-9084.2014.03.007 |
| [8] |
吴建忠, 赵东升, 黄文功 , 等. 12个亚麻品种亲缘关系的SRAP分析. 中国麻业科学, 2012,34(4):153-157.
doi: 10.3969/j.issn.1671-3532.2012.04.001 |
| [9] | Stewart C, Via L . A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques, 1993,14(5):748-750. |
| [10] | Li G, Quiros C F . Sequence-related amplified polymorphism (SRAP),a new marker system based on a simple PCR reaction: its application to mapping and gene tagging in Brassica. Theoretical & Applied Genetics, 2001,103(2-3):455-461. |
| [11] | 孙翊, 张永春, 殷丽青 , 等. 朱顶红SRAP-PCR反应体系的建立和优化. 分子植物育种, 2017,15(5):1806-1813. |
| [12] |
Gresshoff P M . Fast and sensitive silver staining of DNA in polyacrylamide gels. Analytical Biochemistry, 1991,196(1):80-83.
doi: 10.1016/0003-2697(91)90120-I pmid: 1716076 |
| [13] |
张安世, 徐九文, 张利民 , 等. 北方部分粳稻品种遗传关系的SRAP分析. 作物杂志, 2009(6):55-58.
doi: 10.3969/j.issn.1001-7283.2009.06.014 |
| [14] |
石玲艳, 侯建华, 张永虎 , 等. 基于SRAP标记的玉米自交系遗传多样性与群体结构分析. 作物杂志, 2015(3):57-63.
doi: 10.16035/j.issn.1001-7283.2015.03.011 |
| [15] | 李锐, 白建荣, 王秀红 , 等. 144份甜玉米群体的遗传多样性分析. 作物杂志, 2018(2):17-24. |
| [16] | 张帅, 庞玉辉, 王征宏 , 等. 小麦种质资源农艺性状变异及其遗传多样性分析. 作物杂志, 2018(2):44-51. |
| [17] | Botstein D, White R L, Skolnick M , et al. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. American Journal of Human Genetics, 1980,32(3):314-331. |
| [18] |
令狐斌, 侯思宇, 孙朝霞 , 等. 苦荞SRAP分子标记体系优化与遗传多样性分析. 中国农业大学学报, 2015,20(1):37-43.
doi: 10.11841/j.issn.1007-4333.2015.01.05 |
| [19] | 李维卫, 王爱兰, 聂臻臻 . S山茴香RAP反应体系正交设计优化及引物筛选. 基因组学与应用生物学, 2017,36(8):3103-3109. |
| [20] | 延娜, 李巧丽, 郭军战 . 果桑遗传差异的SRAP和EST-SSR分析. 西北农业学报, 2015,24(9):91-97. |
| [21] | 李霞, 李书华, 杨林 , 等. 芦笋种质资源的SRAP遗传多样性分析. 农学学报, 2016,6(3):40-45. |
| [22] | 林春华, 李兆龙, 乔燕春 , 等. SRAP分子标记分析体系的建立及白簕资源亲缘关系分析. 基因组学与应用生物学, 2012,31(5):498-504. |
| [23] | 张良波, 王解香, 李培旺 , 等. 梾木属SRAP-PCR反应体系的优化及引物筛选. 分子植物育种, 2014,12(5):1005-1010. |
| [24] |
杨永, 王豪杰, 张学军 , 等. 新疆甜瓜地方种质资源遗传多样性的SRAP分析. 植物遗传资源学报, 2017,18(3):436-448.
doi: 10.13430/j.cnki.jpgr.2017.03.008 |
| [25] |
黄伟, 鲁敏, 张起 , 等. 利用SRAP标记分析贵州砂梨资源遗传多样性. 西北植物学报, 2016,36(2):280-287.
doi: 10.7606/j.issn.1000-4025.2016.02.0280 |
| [26] | 朱海生, 叶新如, 陈敏氡 , 等. 丝瓜种质资源的SRAP分析. 分子植物育种, 2016,14(8):2217-2223. |
| [1] | Wu Ruixiang,Yang Jianchun,Wang Liqin,Guo Xiujuan. Evaluation of the Adaptability of Flax Drought Resistance Based on Multiple Statistics Analysis [J]. Crops, 2018, 34(5): 10-16. |
| [2] | Ma Mengli,Zheng Yun,Zhou Xiaomei,Zhang Tingting,Zhang Xiaoqian,Lu Bingyue. Genetic Diversity Analysis of Red Rice from Hani’s Terraced Fields in Yunnan Province [J]. Crops, 2018, 34(5): 21-26. |
| [3] | Huiyao Tian,Jizhi Jiang,Chengbin Li,Fen Shen,Ning Hou. Genetic Diversity of Phytophthora infestans in Northeast China [J]. Crops, 2018, 34(3): 168-173. |
| [4] | Shuai Zhang,Yuhui Pang,Zhenghong Wang,Liming Wang,Chunyan Chen,Zhankui Zeng,Chunping Wang. Variation of Agronomic Traits and Genetic Diversity in Wheat Germplasms [J]. Crops, 2018, 34(2): 44-51. |
| [5] | Rui Li,Jianrong Bai,Xiuhong Wang,Congzhuo Zhang,Xiaomei Zhang,Lei Yan,Ruijuan Yang. Population Genetic Diversity of 144 Sweet Maizes [J]. Crops, 2018, 34(2): 17-24. |
| [6] | Lu Zhao,Zhiwei Yang,Liqun Bu,Ling Tian,Mei Su,Lei Tian,Yinxia Zhang,Shuqin Yang,Peifu Li. Analysis and Comprehensive Evalution of Phenotypic Genetic Diversity of Ningxia and Xinjiang Rice Germplasm [J]. Crops, 2018, 34(1): 25-34. |
| [7] | Pengyan Guo,Caiping Wang,Jiecheng Ren,Jiping Zhao,Ying Xu,Maolin Yue. Genetic Diversity of Agronomic Traits of Mung Beans from Different Geographical Sources [J]. Crops, 2017, 33(6): 55-59. |
| [8] | Lianmei Tai,Yuhu Zuo,Yaling Zhang,Yongling Jin,Yongxia Guo,Xuehui Jin,Guanghui Jin. Analysis of Genetic Diversity of Alternaria solani Isolated from Potato in Heilongjiang Province [J]. Crops, 2017, 33(3): 151-156. |
| [9] | Xiujuan Guo,Xuejin Feng,Jianchun Yang,Ruixiang Wu,Liqin Wang. Effects of Interaction between Fertilizer and Density on the Yield and Economic Traits of Flax [J]. Crops, 2017, 33(2): 135-138. |
| [10] | Chaowu Zeng,Xiaodong Liang,Jianjiang Li. Genetic Diversity Analysis on Main Characters of Spring Wheat Germplasms in Central Asia [J]. Crops, 2017, 33(2): 67-71. |
| [11] | Hongxiao Ren,Cuimian Jiang,Yunqing Gao,Qibing Shang,Dongxu Xu,Fang Wang,Xuzhen Cheng. Genetic Diversity and Phenotypic Trait of Chinese Tradional Famous Mungbean [J]. Crops, 2017, 33(1): 44-47. |
| [12] | Hongmei Yuan,Wendong Guo,Lijuan Zhao,Ying Yu,Jianzhong Wu,Lili Cheng,Dongsheng Zhao,Qinghua Kang,Wengong Huang,Yubo Yao,Xixia Song,Weidong Jiang,Yan Liu,Tingfen Ma,Guangwen Wu,Fengzhi Guan. Cloning and Expression Analysis of the Glycosyltransferase Gene LuUGT72E1 in Flax [J]. Crops, 2016, 32(4): 62-67. |
| [13] | Yubo Yao,Guangwen Wu,Wengong Huang,Qinghua Kang,Weidong Jiang,Ying Lu,Shuquan Zhang. Potassium Use Efficiency of Different Flax Genotypes [J]. Crops, 2016, 32(4): 80-85. |
| [14] | Shuyan Wang,Bing Han,Simin Zhou,Jun Xu. Correlation Analysis between the Expression of Stearoyl Acyl Carrier Protein Dehydrogenase Gene and the Soluble Sugar and Fat Content of Oil Flax [J]. Crops, 2016, 32(4): 56-61. |
| [15] | Xiuxia Cao,Aiping Qian,Wei Zhang,Chongqing Yang. The Effects of Different Amounts of Zinc Fertilizer on the Growth and Yield of Oil-Flax in Arid Land [J]. Crops, 2016, 32(3): 167-170. |
|
||