Crops ›› 2019, Vol. 35 ›› Issue (6): 33-42.doi: 10.16035/j.issn.1001-7283.2019.06.006
Previous Articles Next Articles
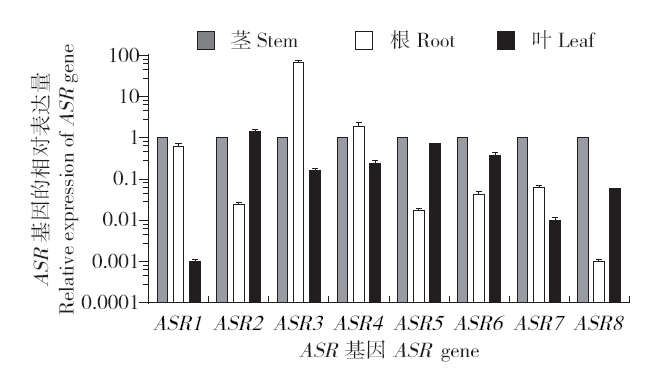
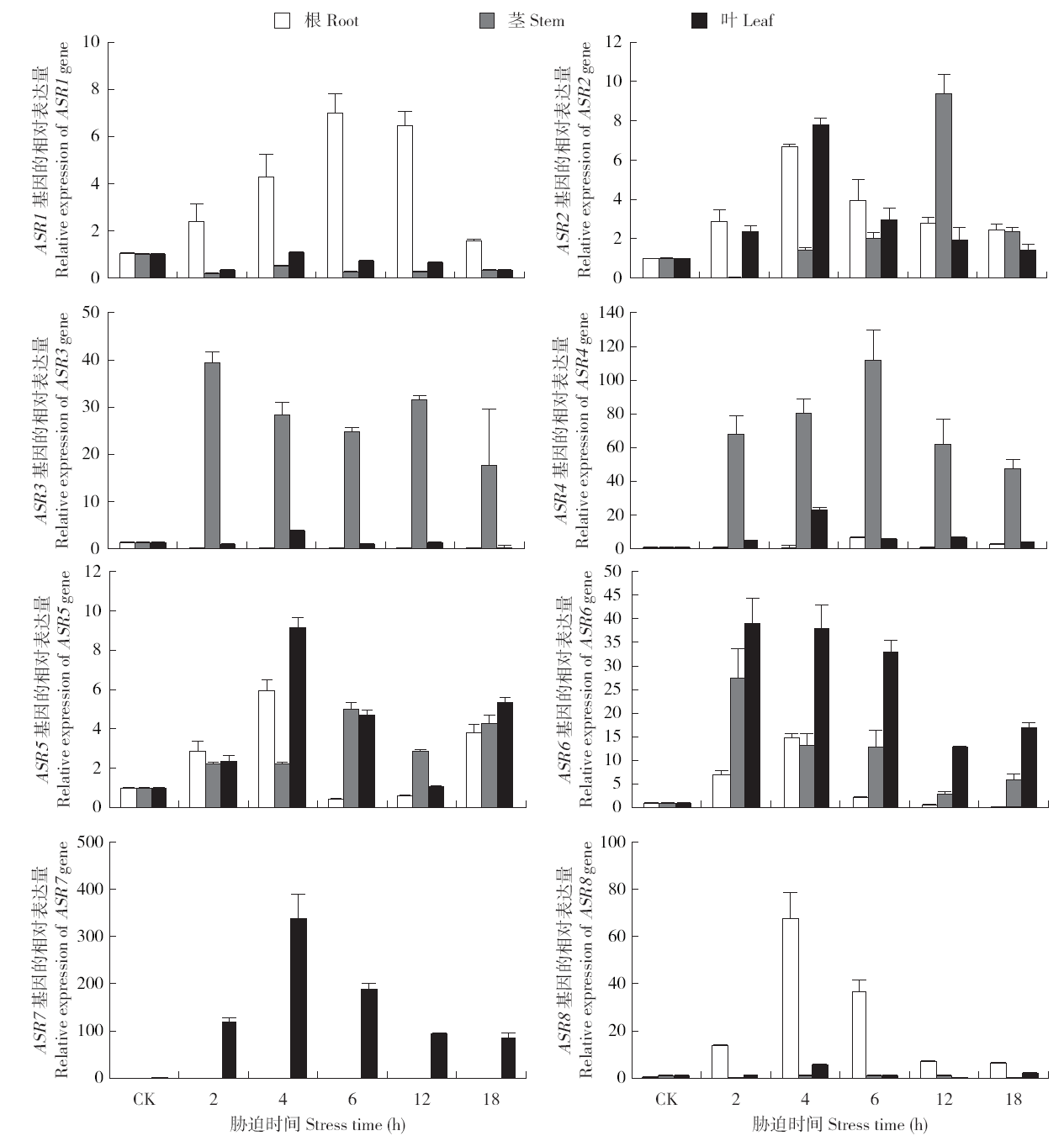
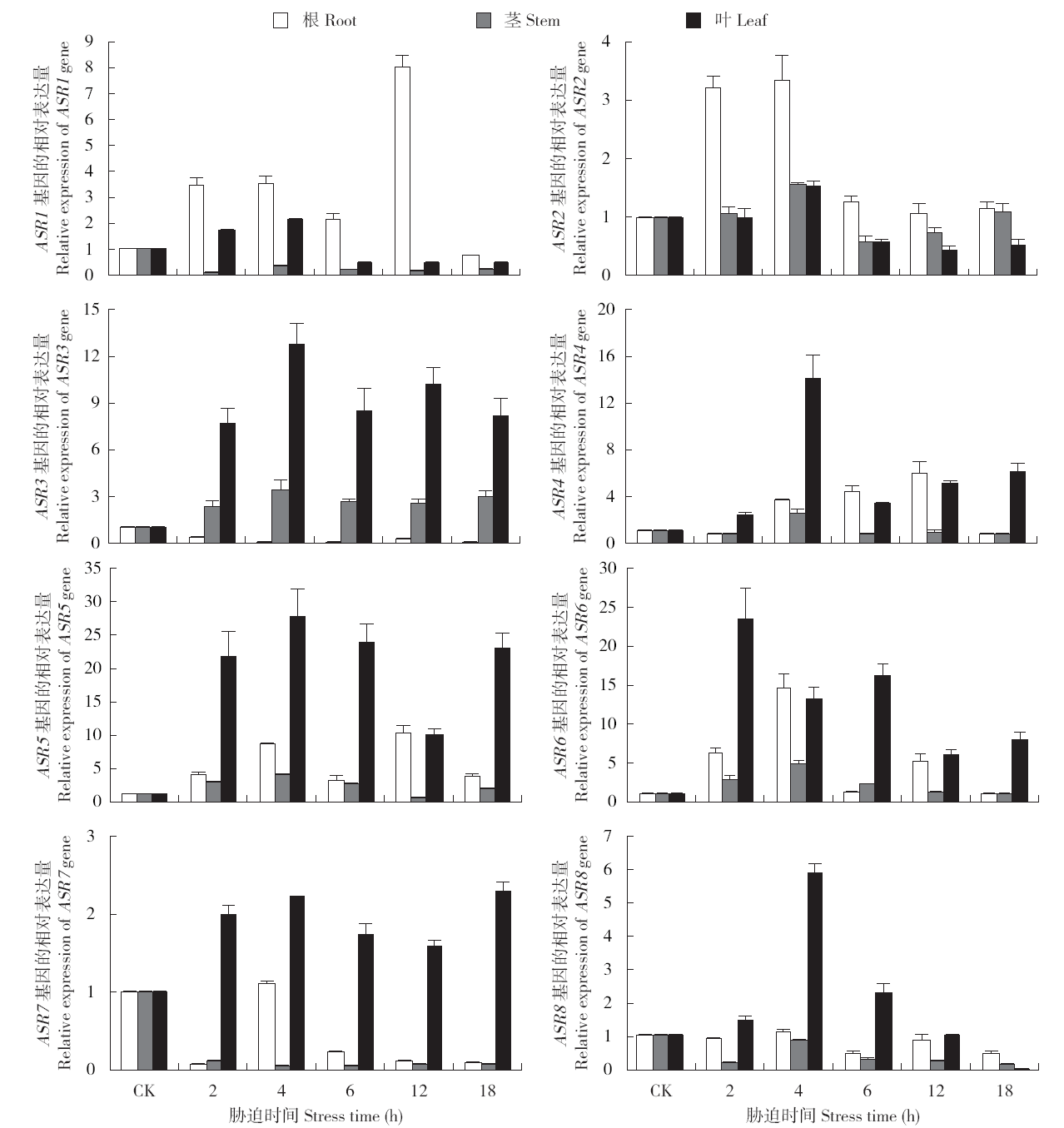
Identification and Expression Analysis of ASR Family Genes in Setaria italica
Song Jian1,2,Cao Xiaoning3,Wang Haigang3,Chen Ling3,Wang Junjie3,Liu Sichen3,Qiao Zhijun3
- 1College of Biological Engineering, Shanxi University, Taiyuan 030006, Shanxi, China
2Institute of Industrial Crop, Shanxi Academy of Agricultural Sciences, Fenyang 032200, Shanxi, China
3Institute of Crop Germplasm Resources, Shanxi Academy of Agricultural Sciences, Taiyuan 030031, Shanxi, China
| [1] | 海宝, 双权, 松梅 . 优质饲草料—谷子. 内蒙古草业,1998(4):10. |
| [2] | 刘敬科, 刁现民 . 我国谷子产业现状与加工发展方向. 农业工程技术(农产品加工业),2013(12):15-17. |
| [3] | 李霞 . 当前山西省谷子品种区试、审(认)定与推广概况. 中国种业,2014(7):9-10. |
| [4] | 张书敏, 刘红云, 程金金 , 等. 快速徒手切片法观察谷子和水稻叶片显微结构. 基因组学与应用生物学, 2015,34(7):1527-1530. |
| [5] | 贾冠清, 刁现民 . 谷子[Setaria italica (L.) P. Beauv.]作为功能基因组研究模式植物的发展现状及趋势. 生命科学, 2017,29(3):292-301. |
| [6] | 张瑞杰, 王喆, 连卜颖 , 等. 谷子ABC转运蛋白基因与抗旱关系的研究. 山西农业大学学报(自然科学版), 2018,38(1):11-15. |
| [7] | 杨晔, 李晶, 顾万荣 , 等. Asr基因家族的研究进展. 作物杂志,2013(3):7-11. |
| [8] | 张丽丽, 李博, 谭燕华 , 等. 玉米转录因子基因ZmASR1的克隆及植物表达载体构建. 江苏农业科学, 2016,44(4):16-22. |
| [9] |
Hagit A Z, Scolnik P A, Dudy B Z . Tomato Asr1 mRNA and protein are transiently expressed following salt stress,osmotic stress and treatment with abscisic acid. Plant Science, 1995,110(2):205-213.
doi: 10.1016/0168-9452(95)94515-K |
| [10] |
Joo J, Lee Y H, Choi D H , et al. Rice ASR1 has function in abiotic stress tolerance during early growth stages of rice. Journal of the Korean Society for Applied Biological Chemistry, 2013,56(3):349-352.
doi: 10.1007/s13765-013-3060-6 |
| [11] |
Yang C Y, Chen Y C, Guang Y J , et al. A lily asr protein involves abscisic acid signaling and confers drought and salt resistance in Arabidopsis. Plant Physiology, 2005,139(2):836-846.
doi: 10.1104/pp.105.065458 pmid: 16169963 |
| [12] |
Wang J T, Gould J H, Padmanabhan V , et al. Analysis and localization of the Water-Deficit Stress-Induced gene (lp3). Journal of Plant Growth Regulation, 2002,21(4):469-478.
doi: 10.1007/s00344-002-0128-7 |
| [13] |
Jha B, Lal S, Tiwari V , et al. The SbASR-1 gene cloned from an extreme halophyte Salicornia brachiata enhances salt tolerance in transgenic tobacco. Marine Biotechnology, 2012,14(6):782-792.
doi: 10.1007/s10126-012-9442-7 |
| [14] |
Feng Z J, Xu Z S, Sun J T , et al. Investigation of the ASR family in foxtail millet and the role of ASR1 in drought/oxidative stress tolerance. Plant Cell Reports, 2016,35(1):115-128.
doi: 10.1007/s00299-015-1873-y pmid: 26441057 |
| [15] | 李建锐 . 谷子SiASR4基因参与植物响应干旱和盐胁迫的功能研究. 北京:中国农业大学, 2018. |
| [16] |
Liu Y, Teng X, Yang X , et al. Shotgun proteomics and network analysis between plasma membrane and extracellular matrix proteins from rat olfactory ensheathing cells. Cell Transplantation, 2010,19(2):133-146.
doi: 10.3727/096368910X492607 pmid: 20350363 |
| [17] | 刘晓庆, 陈华涛, 张红梅 , 等. 大豆GmbHLH041基因的生物信息学分析及对镉胁迫的响应. 华北农学报, 2018,33(3):14-18. |
| [18] |
Lu Y, Liu L, Wang X , et al. Genome-wide identification and expression analysis of the expansin gene family in tomato. Molecular Genetics and Genomics, 2016,291(2):597-608.
doi: 10.1007/s00438-015-1133-4 pmid: 26499956 |
| [19] | 邢国芳, 张莉, 冯万军 , 等. 谷子WRKY 转录因子基因家族分析. 山西农业大学学报(自然科学版), 2016,36(12):837-845. |
| [20] | 孙洋洋 . Boule基因的生物信息学分析. 呼和浩特:内蒙古师范大学, 2014. |
| [21] |
Lescot M, DeHais P, Thijs G ,et al. Plant CARE,a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research, 2002,30(1):325-327.
doi: 10.1093/nar/30.1.325 pmid: 11752327 |
| [22] |
高堃, 华营鹏, 宋海星 , 等. 甘蓝型油菜PIN家族基因的鉴定与生物信息学分析. 作物学报, 2018,44(9):1334-1346.
doi: 10.3724/SP.J.1006.2018.01334 |
| [1] | Yue Linqi,Shi Weiping,Guo Jiahui,Guo Pingyi,Guo Jie. Response of Cutin Synthetic Genes of Foxtail Millet to Drought Stress [J]. Crops, 2019, 35(4): 183-190. |
| [2] | Lü Liangjie,Chen Xiyong,Zhang Yelun,Liu Qian,Wang Limei,Ma Le,Li Hui. Bioinformatics Identification of GASA Gene Family Expression Profiles in Wheat [J]. Crops, 2018, 34(6): 58-67. |
| [3] | Haibin Luo, Shengli Jiang, Chengmei Huang, Huiqing Cao, Zhinian Deng, Kaichao Wu, Lin Xu, Zhen Lu, Yuanwen Wei. Cloning and Expression of ScHAK10 Gene in Sugarcane [J]. Crops, 2018, 34(4): 53-61. |
| [4] | Ying Zhang,Pengyu Liu,Xue Bai,Yang Yang,Yueying Li. Expression and Bioinformatics Analysis of CsWRKY23 Gene in Cucumber [J]. Crops, 2017, 33(5): 38-42. |
| [5] | Hongmei Yuan,Wendong Guo,Lijuan Zhao,Ying Yu,Jianzhong Wu,Lili Cheng,Dongsheng Zhao,Qinghua Kang,Wengong Huang,Yubo Yao,Xixia Song,Weidong Jiang,Yan Liu,Tingfen Ma,Guangwen Wu,Fengzhi Guan. Cloning and Expression Analysis of the Glycosyltransferase Gene LuUGT72E1 in Flax [J]. Crops, 2016, 32(4): 62-67. |
| [6] | Yu Li,Wenyin Zheng,Chun Feng,Rongfu Wang,Juan Li. Analysis of FeSOD Gene Expression in Normal Wheat Yannong 19 Seedlings under Abiotic Stress [J]. Crops, 2016, 32(4): 75-79. |
| [7] | . [J]. Crops, 2013, 29(3): 7-11. |
|