Crops ›› 2024, Vol. 40 ›› Issue (1): 65-72.doi: 10.16035/j.issn.1001-7283.2024.01.009
Previous Articles Next Articles
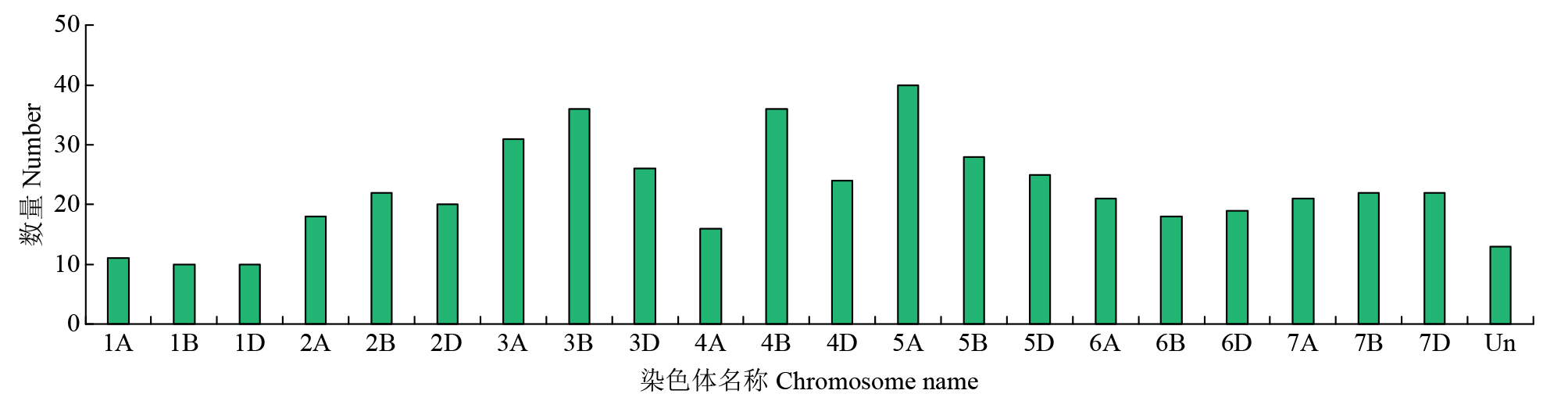
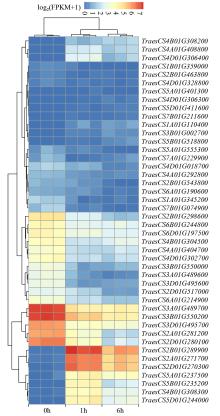
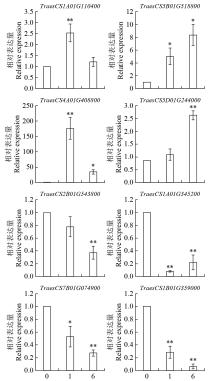
Identification of bHLH Family Transcription Factors of Wheat and Expression Analysis under Salt Stress
Lü Baolian1,2( ), Yang Yuxin2, Cui Licao2, Shi Feng3, Ma Liang3, Kong Xiuying2, Zhang Lichao2(
), Yang Yuxin2, Cui Licao2, Shi Feng3, Ma Liang3, Kong Xiuying2, Zhang Lichao2( ), Ni Zhiyong1(
), Ni Zhiyong1( )
)
- 1College of Life Sciences, Xinjiang Agricultural University, Urumqi 830052, Xinjiang, China
2Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100081, China
3Shijiazhuang Academy of Agricultural and Forestry Sciences, Shijiazhuang 050041, Hebei, China
| [1] |
Zhang H M, Zhu J H, Gong Z Z, et al. Abiotic stress responses in plants. Nature Reviews Genetics, 2022, 23(2):104-119.
doi: 10.1038/s41576-021-00413-0 |
| [2] |
Bhanbhro N, Xiao B B, Han L, et al. Adaptive strategy of allohexaploid wheat to long-term salinity stress. BMC Plant Biology, 2020, 20(1):210.
doi: 10.1186/s12870-020-02423-2 pmid: 32397960 |
| [3] |
马彦军, 段慧荣, 魏佳, 等. NaCl胁迫下黑果枸杞转录组测序分析. 生物技术通报, 2020, 36(2):100-109.
doi: 10.13560/j.cnki.biotech.bull.1985.2019-0492 |
| [4] | 朱涛, 李芳菲, 杨海涵, 等. 山药bHLH基因家族鉴定及表达分析. 信阳师范学院学报(自然科学版), 2022, 35(3):393-399. |
| [5] |
Hao Y Q, Zong X M, Ren P, et al. Basic helix-loop-helix (bHLH) transcription factors regulate a wide range of functions in Arabidopsis. International Journal of Molecular Sciences, 2021, 22(13):7152.
doi: 10.3390/ijms22137152 |
| [6] | Wang Y G, Li H Y, University H, et al. Sugar beet BvBHLH 92 tissue expression and subcellular localization. Journal of Engineering of Heilongjiang University, 2017, 8(3):45-49,2. |
| [7] | Lee H Y, Seo J S, Um T Y.OsbHLH148 confers drought tolerance in Arabidopsis. International Plant and Animal Genome Conference, January 14-18, 2012. California: San Diego, 2012. |
| [8] | 孙玉合, 孙晋浩, 牛文利, 等. 烟草NtbHLH112基因的克隆、鉴定及表达模式分析. 中国烟草科学, 2020, 41(5):8-14. |
| [9] |
悦曼芳, 张春, 郑登俞, 等. 玉米转录因子ZmbHLH91对非生物逆境胁迫的应答. 作物学报, 2022, 48(12):3004-3017.
doi: 10.3724/SP.J.1006.2022.13060 |
| [10] |
Zhai Y Q, Zhang L C, Xia C, et al. The wheat transcription factor, TabHLH39, improves tolerance to multiple abiotic stressors in transgenic plants. Biochemical and Biophysical Research Communications, 2016, 473(4):1321-1327.
doi: S0006-291X(16)30577-0 pmid: 27091431 |
| [11] |
Wang F B, Zhu H, Kong W L, et al. The Antirrhinum AmDEL gene enhances flavonoids accumulation and salt and drought tolerance in transgenic Arabidopsis. Planta, 2016, 244(1):59-73.
doi: 10.1007/s00425-016-2489-3 |
| [12] |
Sun X, Wang Y, Sui N. Transcriptional regulation of bHLH during plant response to stress. Biochemical and Biophysical Research Communications, 2018, 503(2):397-401.
doi: S0006-291X(18)31625-5 pmid: 30057319 |
| [13] | 毕晨曦, 杨宇昕, 于月华, 等. 小麦bZIP家族转录因子的鉴定及其在盐胁迫条件下的表达分析. 分子植物育种, 2021, 19 (15):4887-4895. |
| [14] |
Gabriela T, Enamul H, Peter H Q. The Arabidopsis basic/ helix-loop-helix transcription factor family. Plant Cell, 2003, 15:1749-1770.
doi: 10.1105/tpc.013839 |
| [15] |
Lorenzo C P, Anahit G, Irma R V, et al. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiology, 2010, 153(3):1398-1412.
doi: 10.1104/pp.110.153593 |
| [16] | 赵小波, 闫彩霞, 李春娟, 等. 花生转录因子AhbHLH18克隆与功能分析. 花生学报, 2022, 51(2):1-8. |
| [17] | 施田野, 顾宇蓝, 张磊, 等. 粗山羊草响应盐胁迫转录组分析. 分子植物育种, 2020, 18(21):7015-7022. |
| [18] | 田烨, 王爽, 路正禹, 等. 甜菜应答盐胁迫诱导表达bHLH基因的鉴定与分析. 黑龙江大学自然科学学报, 2020, 37(6):712-717. |
| [19] | 徐秀荣, 杨克彬, 王思宁, 等. 毛竹bHLH转录因子的鉴定及其在干旱和盐胁迫条件下的表达分析. 植物科学学报, 2019, 37(5):610-620. |
| [20] | 唐文武, 吴秀兰, 钟佩桥, 等. 白菜bHLH转录因子家族的全基因鉴定及表达特征分析. 江西农业学报, 2020, 32(6):1-5. |
| [21] | 黄小芳, 毕楚韵, 王和寿, 等. 甘薯基因组bHLH转录因子鉴定与逆境胁迫表达分析. 福建农林大学学报(自然科学版), 2021, 50(4):440-450. |
| [22] | 孙颖琦, 孟亚轩, 赵心月, 等. 谷子bHLH转录因子家族基因鉴定及生物信息学分析. 种子, 2021, 40(12):45-55. |
| [23] |
何洁, 顾秀容, 魏春华, 等. 西瓜bHLH转录因子家族基因的鉴定及其在非生物胁迫下的表达分析. 园艺学报, 2016, 43(2):281-294.
doi: 10.16420/j.issn.0513-353x.2015-0886 |
| [1] | Liu Hongjie, Ren Dechao, Ge Jun, Zhang Suyu, Lü Guohua, He Xun. Effects of Accumulated Temperature and Planting Density on Pre-Winter Growth of Wheat [J]. Crops, 2024, 40(1): 141-147. |
| [2] | Liu Zhewen, Guo Dandan, Chang Xuhong, Wang Demei, Yang Yushuang, Liu Xiwei, Wang Yujiao, Shi Shubing, Wang Yanjie, Zhao Guangcai. Effects of Nitrogen Dressing Time and Proportion on Wheat Grain Filling and Its Physiological Mechanism [J]. Crops, 2024, 40(1): 174-179. |
| [3] | Hao Xiaocong, Li Xinyu, Hou Qiling, Yang Jifang, An Chunhui, Wang Changhua, Ye Zhijie, Zhang Fengting. Effects of Nitrogen Application Rate on the Quality of Two-Line Hybrid Wheat [J]. Crops, 2024, 40(1): 187-192. |
| [4] | Hao Yani, Pei Hongbin, Gao Zhenfeng, Zhang Yijun, Yang Zhenping. Effects of Bacillus vallismortis and Straw Replacing Phosphorus Fertilizer on Growth, Yield and Quality of Tartary Buckwheat [J]. Crops, 2024, 40(1): 204-213. |
| [5] | Zhang Rong, Jiang Enxi, Chen Si, Yu Xurun, Chen Gang, Ran Liping, Xiong Fei. Study on the Grain Formation in Wheat Spike Regulated by Ethephon and 1-Methylcyclopropene [J]. Crops, 2023, 39(6): 101-107. |
| [6] | Liu Zhewen, Guo Dandan, Chang Xuhong, Wang Demei, Wang Yanjie, Yang Yushuang, Liu Xiwei, Wang Yujiao, Shi Shubing, Zhao Guangcai. Response of Nitrogen Accumulation and Translocation after Anthesis in Strong Gluten Wheat to Nitrogen Topdressing Period and Proportion [J]. Crops, 2023, 39(6): 114-120. |
| [7] | Liu Xiwei, Wang Demei, Wang Yanjie, Yang Yushuang, Zhao Guangcai, Chang Xuhong. Impacts Mechanism of Drought and Heat Stress in the Middle and Late Growing Period on Wheat Grain Yield Formation Process and Mitigation Measures [J]. Crops, 2023, 39(6): 17-25. |
| [8] | Chen Dan, Xiong Furong, Wu Shaoyun, Bai Xiaodong, Zhou Guoyan, Wu Xiaoyang, Cai Qing. Molecular Detection and Geographic Distribution of Stripe Rust Resistance Gene Loci in Yunnan Wheat Landraces [J]. Crops, 2023, 39(6): 41-46. |
| [9] | Wang Yifan, Ren Ning, Dong Xiangyang, Zhao Yanan, Ye Youliang, Wang Yang, Huang Yufang. Effects of Controlled-Release and Ordinary Urea on Wheat Yield, Nitrogen Absorption and Economic Benefit [J]. Crops, 2023, 39(5): 117-123. |
| [10] | Yang Mei, Yang Weijun, Gao Wencui, Jia Yonghong, Zhang Jinshan. Effects of Combined Application of Biochar and Nitrogen Fertilizer on Dry Matter Transport, Agronomic Characteristics and Yield of Winter Wheat in Irrigation Area [J]. Crops, 2023, 39(5): 138-144. |
| [11] | Huang Jie, Ge Changbin, Wang Jun, Cao Yanyan, Qiao Jiliang, Liao Pingʼan, Song Danyang, Lu Wenying. Simulation Model of Relative Meteorological 1000-Grain Weight of Wheat of Luohe Based on Principal Component Regression [J]. Crops, 2023, 39(5): 212-218. |
| [12] | Liu Shuhan, Chen Lei, Zhang Jianchao, Hu Gan, Sun Junyan, Liu Dongtao, Wang Junwei. Gene Differential Expression Analysis of TMS5 in the Fertility Conversion of Wheat BNS Sterile Line [J]. Crops, 2023, 39(5): 24-29. |
| [13] | Zhang Dongxu, Hu Danzhu, Yan Jinlong, Feng Liyun, Wu Zhiyuan, Zhang Junling, Li Yanhua. Effects of Spraying Streptomyces on Yield and Photosynthetic Characteristics of Late-Sown Wheat under Different Crop Rotations [J]. Crops, 2023, 39(5): 255-263. |
| [14] | Song Guicheng, Yu Guihong, Zhang Peng, Ma Hongxiang. Evaluation on Waterlogging Resistance of Different Wheat Varieties (Lines) at Jointing Stage [J]. Crops, 2023, 39(5): 30-36. |
| [15] | Ge Changbin, Qin Suyan, Qiao Jiliang, Wang Jun, Qi Shuangli, Lu Wenying, Zhang Zhenyong. Comparative Analysis of Agronomic Traits, Quality and Disease Evolution of Approved Wheat Varieties in Southern Henan and Southern Huai River in Jiangsu from 2001 to 2021 [J]. Crops, 2023, 39(5): 49-58. |
|
||