开放科学(资源服务)标识码(OSID): 
类黄酮类化合物是莽草酸酯和苯丙酯途径产生的苯丙类衍生物[15]。苯丙氨酸的生物合成始于脱氨酶(PAL)的脱氨基作用,其次是反应催化肉桂酸-4-羟化酶(C4H)、4-香豆酸辅酶A连接酶(4CL)、查耳酮合酶(CHS)、查耳酮异构酶(CHI)、黄酮3-羟化酶(F3H)、类黄酮3-羟化酶(F3ʹH)、黄酮醇合成酶(FLS)及糖基转移酶(GTs)等酶的共同作用与参与[16,17]。目前,关于苦荞黄酮类合成相关酶基因的调控机理尚不清楚,尤其是关于不同黄酮类物质之间的转化关系(糖基化作用)的研究还有所欠缺。本文主要研究了1个苦荞黄酮合成途径中的酶基因FtF3GT1,探究其在黄酮合成途径中的作用,为解析苦荞的黄酮合成机理提供基础。
1 材料与方法
1.1 苦荞材料及种植培养
本试验采用的川荞1号由中国农业科学院作物科学研究所荞麦基因资源创新研究组提供。试验于2019年在中国农业科学院作物科学研究所荞麦基因资源创新研究组实验室完成。将川荞1号种子用1%次氯酸钠溶液消毒6min;然后用75%乙醇灭菌2min;再用无菌水清洗直至水澄清为止。将种子放在灭菌的滤纸上吸干水分,种植在MS培养基上。于22℃~25℃、光周期16h/8h、湿度75%~80%的条件下培养。
1.2 RNA提取与cDNA合成
选取14d幼苗50~100mg,加液氮充分研磨后,采用Trizol法[18]提取总RNA,以此RNA为模板,使用HiScript® III 1st Strand cDNA Synthesis Kit (+gDNA wiper)试剂盒(南京诺唯赞生物科技有限公司)进行反转录,获得川荞1号的cDNA。
1.3 FtF3GT1基因CDS的克隆与序列分析
在中国农业科学院作物科学研究所荞麦基因资源创新研究组提供MeJA处理的苦荞转录组数据库中筛选获得FtF3GT1基因,根据FtF3GT1基因的ORF设计特异引物(表1),以川荞1号cDNA为模板,以1.2所述DNA为模板进行PCR扩增,获得目的基因的CDS序列。PCR程序为95℃预加热3min;95℃保持30s使模板变性;60℃复性30s,使引物和模板充分退火,72℃保持60s,使引物充分延伸,合成DNA,完成一个循环,共35个循环。PCR纯化产物连接到pTOPO-Blunt Simple平末端克隆载体上,获得FtF3GT1-T载体质粒。由北京博迈德生物技术有限公司测序、分析和拼接后得到FtF3GT1基因全长序列。将FtF3GT1基因氨基酸序列在NCBI数据库中Blast比对,获得其他物种GTs类蛋白序列。采用Bioxm软件翻译氨基酸序列,预测其蛋白质大小及等电点,利用MEGA 7.0软件中的Neighbour-Joining算法构建系统进化树。
表1 引物序列
Table 1
| 引物名称Primer name | 引物序列(5'-3') Primer sequence (5'-3') |
|---|---|
| FtF3GT1-F(CDS) | 5'-ATGGGAACCCAATCAAGC-3' |
| FtF3GT1-R(CDS) | 5'-CTACTGCTTACCAACCAAAC-3' |
| pCAMBIA1307-FtF3GT1-F | 5'-GGGGGCGGCCGCTCTAGAATGGGAACCCAATCAAGC-3' |
| pCAMBIA1307-FtF3GT1-R | 5'-AAGCTTGATATCGAATTCCTACTGCTTACCAACCAAAC-3' |
| pCAMBIA3301-FtF3GT1-F | 5'-GAATCCTTAAATGGTTCATATTACTATGCAATTAG-3' |
| pCAMBIA3301-FtF3GT1-R | 5'-TCTAGATTTGTAAAATGCTTGATTTGTTTCTTG-3' |
| FtH3-QF | 5'-GAAATTCGCAAGTACCAGAAGAG-3' |
| FtH3-QR | 5'-CCAACAAGGTATGCCTCAGC-3' |
| FtF3GT1-QF | 5'-TCAAATAAGCTCGCCTC-3' |
| FtF3GT1-QR | 5'-GCTGCATTTTGTCAAGAGCG-3' |
| FtActin-QF | 5'-GAAATTCGCAAGTACCAGAAGAG-3' |
| FtActin-QR | 5'-CCAACAAGGTATGCCTCAGC-3' |
1.4 FtF3GT1基因的表达分析
1.4.1 FtF3GT1基因的时空表达分析 提取苦荞麦刚长出真叶时(7d左右)的根、茎、叶、成熟种子和未成熟种子的总RNA,RNA提取方法同1.2。
1.4.2 不同激素诱导FtF3GT1基因表达分析 无菌苗在组培室光照培养至刚长出真叶时(7d左右),取3株长势一致的无菌苗于液体MS培养基中,室温震荡(120转/min)处理1d后用MeJA(50µmol/L)处理1、4和12h时后取样(作为样品)。处理其他条件和培养均保持一致,对照组添加同体积的二甲基亚砜(DMSO)。取样之后用灭菌滤纸快速吸干水分后置于液氮中速冻,于-80°C冰箱中保存备用,RNA提取方法同1.2。
1.4.3 实时荧光定量PCR 以荞麦组成型表达的FeH3F基因为内参,引物名称为FtH3-QF/R。同时设计基因特异性引物FtF3GT1-QF/R(表1),以1.2中制备好的cDNA为模板,作3次生物学重复试验,利用实时荧光定量PCR(quantitative real-time PCR,qRT-PCR)在BAI 7500实时荧光定量PCR仪上检测FtF3GT1基因表达量。qPCR反应体系同谈天斌等[19]所述,本试验中用RE(相对表达量)=2-ΔΔCT的算法,计算目标基因的相对表达量;设置诱导表达control 2-ΔΔCT值为1,计算出MeJA(50µmol/L)分别处理1、4和12h的表达倍数。
1.5 农杆菌介导的苦荞毛状根转化系统的构建与遗传转化
1.5.1 植物表达载体构建 pCAMBIA1307-FtF3GT1过表达载体的构建。设计含XbaI和KpnI酶切位点的同源重组引物,以FtF3GT1-T载体为模板,pCAMBIA1307-FtF3GT1-F/R为引物(表1),PCR扩增FtF3GT1基因的全长序列。随后经酶切、回收和连接转化后,将FtF3GT1基因全长序列正向插入pCAMBIA-1307载体的CaMV35S启动子后,经北京博迈德生物技术有限公司测序完全后得到过表达载体pCAMBIA1307-FtF3GT1。
pCAMBIA3301:FtF3GT1::GUS表达载体的构建。为了进一步探究MeJA对FtF3GT1基因表达的影响,根据FtF3GT1基因的启动子序列,设计含XbaI和EcoRI酶切位点的特异引物(表1),以川荞1号的DNA为模板,pCAMBIA3301-FtF3GT1-F/R为引物,克隆了FtF3GT1基因上游1 200bp的启动子序列,并连接到pTOPO-Blunt Simple平末端克隆载体上,经测序后得到pCAMBIA3301-FtF3GT1-T载体质粒。随后经酶切、回收和连接转化后,将FtF3GT1基因启动子序列连接到pCAMBIA3301表达载体上。经测序完全后得到表达载体pCAMBIA3301:FtF3GT1pro::GUS。
1.5.2 农杆菌介导的苦荞毛状根转化 分别吸取2mL A4菌液加到100mL YEB(含Rif 100mg/mL)液体培养基中,在28℃,200转/min摇床中培养12h,菌液浓度OD600=0.8;随后将菌液离心,得到的菌体使用20mL MS液体培养基吹打混匀,重新悬浮。用剪刀剪取2周龄大小的苦荞无菌苗,取子叶和下胚轴切成小段,放入准备好的A4发根农杆菌的菌液中进行侵染,外植体接触菌液时间为15min,轻轻摇动菌体悬浮液,用镊子取出外植体,采用灭菌滤纸吸去外植体上多余的菌体,然后将外植体平铺于垫有两层无菌滤纸的MS固体培养基上。放置在22℃温室中共培养2d后,将外植体取出用无菌滤纸吸干菌液,然后再将外植体放入MS(含100mg/mL头孢霉素)固体培养基中培养[20]。
7~14d后外植体分化出毛状根,选取生长状态良好的毛状根(8~10cm)移入MS(含100mg/mL头孢霉素)固体培养基中培养,22℃光下培养。剪切5cm长度的毛状根用CTAB法提取DNA并鉴定,得到的阳性毛状根即可进行下一步试验。
1.5.3 GUS染色试验 剪切8cm左右的pCAMBIA3301:FtF3GT1pro::GUS阳性毛状根于MS固体培养基上培养,待毛状根的量足够后,取适量置于MS液体培养基中,室温震荡(120转/min)处理同时,转pCAMBIA3301空载体阳性毛状根作为阳性对照,转A4毛状根作为阴性对照。24h后用MeJA(50µmol/L)分别处理1、4和12h时后取样,其他条件和培养均保持一致。取样之后用GUS缓冲液冲洗2遍,再用(GUS缓冲液+50mg/mL X-Gluc)过夜染色(样品放于37℃条件下),待肉眼看得到明显的蓝色后,用75%乙醇脱色完全后拍照。
1.5.4 转基因毛状根的总黄酮含量分析 总黄酮的提取条件为超声25min、温度为50℃,超声频率为40kHz。用80%甲醇溶液提取毛状根中的总黄酮,采用三氯化铝法[21]测定转基因苦荞毛状根的总黄酮含量,期间不间断摇晃试管。在波长为420nm处测定吸光度,同时绘制芦丁标准曲线,以吸光度值A为纵坐标,芦丁浓度(mg/mL)为横坐标,最后用芦丁标准曲线方程计算总黄酮含量。
2 结果与分析
2.1 FtF3GT1基因CDS和启动子的克隆
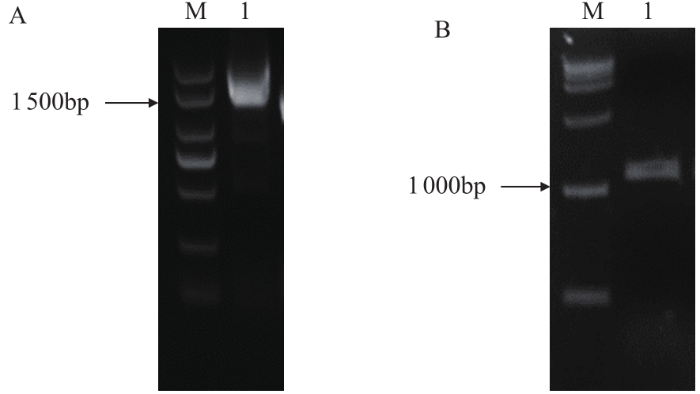
分别以川荞1号的cDNA和DNA为模板,克隆得到FtF3GT1基因的CDS和启动子序列(图1),通过DNAMAN软件进行比对,所获得的CDS、启动子序列与转录组测序筛选到的序列基本一致。
图1
图1
FtF3GT1基因CDS和启动子的克隆
A:M:DL 2 000 DNA标记;1:FtF3GT1基因CDS扩增产物;B:M:DL 2 000 DNA标记;1:FtF3GT1基因启动子的扩增产物
Fig.1
Cloning of CDS and promoter of FtF3GT1 gene
A: M: DL 2 000 DNA marker; 1: PCR product of FtF3GT1 gene CDS; B: M:DL 2 000 DNA marker; 1: PCR product of FtF3GT1 gene promoter
2.2 蛋白和启动子序列分析
氨基酸序列分析发现,该基因的CDS长度为1 401bp,编码蛋白467个氨基酸,分子量51.84kDa,所编码蛋白的等电点为6.5。在NCBI数据库中用Blast比对发现,该蛋白与甜荞麦中的鼠李糖基转移酶FeF3G6″RhaT同源性最高(96.17%),但目前没有对其功能进行深入研究,鉴于其同源蛋白是以黄酮醇3-O-糖苷(1->6)鼠李糖基转移酶为供体,暂时将其命名为FtF3GT1。用NCBI的CD-search在线分析FtF3GT1基因编码氨基酸的保守域,发现所编码的蛋白属于GT-B型糖基转移酶,序列比对发现,FtF3GT1与其他GTs一样,在C端存在一段HAGFGS氨基酸序列,类似于高度保守的氨基酸序列HCGWNS,序列上存在某些氨基酸的突变,猜测这些突变位点可能是黄酮合成相关的关键位点,后续需要更多的试验证据来证明(图2)。
图2
图2
FtF3GT1与其他植物GTs蛋白序列多重比对
Fig.2
Multi-alignment of FtF3GT1 with GTs type proteins from other plants
PhUF5GT (Petunia hybrida BAA89009); AtUF7GT (Arabidopsis thaliana NP567955); FtUF7GT (Fagopyrum esculentum BBF24939); FtUF7GT (F.esculentum BBF24939); FtUFGT2 (F.tataricum KX216513); FtUFGT3 (F. tataricum KX216514); GeUF7GT (Glycyrrhiza echinata BAC784380); GhUF5GT (Glandularia hybrida AB013598); GtUF3GT (Gentiana triflora BAA12737); HvUF3GT (Hordeum vulgare X15694); IhUF5GT (Iris hollandica AB113664); LbUF7GT (Lycium barbarum BAG80536); NtUF7GT (Nicotiana tabacum AAB36653); PyUF3GT (Petunia hybrida BAA89008); SbUF7GT (Scutellaria baicalensis BAA83484); ThUF5GT (Torenia hybrid AB076698); VmUF3GT (Vigna mungo BAA36972); VvUF3GT (Vitis vinifera AAB81683)
将FtF3GT1与其他物种中GTs类蛋白序列进行比对,采用MEGA 7.0软件对这些蛋白通过邻接法构建进化树,并进行分子发育分析发现(图3),FtF3GT1与UG7T型糖基转移酶聚为一类,且与甜荞中的FeUF7GT蛋白亲缘关系最近。
图3
图3
FtF3GT1与其他植物GTs蛋白氨基酸序列的系统发育分析
Fig.3
Phylogenetic analysis of amino acid sequences of FtF3GT1 and GTs from other plants
根据PlantCare生物信息学分析发现(表2),FtF3GT1基因启动子序列中除了包含大量的TATA-box和CAAT-box核心启动子元件,还有其他几类顺式反应元件:G-box、GARE-motif、ABRE、TGACG-motif、TCA-element和TCA-element等。
表2 FtF3GT1基因启动子序列中的顺式作用元件
Table 2
| 位点名称Site name | 序列Sequence | 位点功能Function of site |
|---|---|---|
| TGACG-motif | TGACG | 参与MeJA反应的顺式作用调控元件cis-acting regulatory element involved in the MeJA-responsiveness |
| CGTCA-motif | CGTCA | 参与MeJA反应的顺式作用调控元件cis-acting regulatory element involved in the MeJA-responsiveness |
| ABRE | ACGTG | 参与ABA反应的顺式作用调控元件cis-acting element involved in the abscisic acid responsiveness |
| GARE-motif | TCTGTTG | 响应GA元素gibberellin-responsiveness |
| P-box | CCTTTTG | 响应GA元素gibberellin-responsiveness |
| TCA-element | CCATCTTTTT | 参与SA反应的顺式作用调控元件cis-acting element involved in salicylic acid responsiveness |
| G-box | CACGTC | 光调控元件cis-acting regulatory element involved in light responsiveness |
| ARE | AAACCA | 厌氧诱导所必需的顺式调节元件cis-acting regulatory element essential for the anaerobic induction |
| TATA-box | TATATA | 转录起始-3-O核心启动子元件core promoter element of 3-O transcription start |
2.3 FtF3GT1基因的表达特性
2.3.1 FtF3GT1基因的组织特异性表达 qRT-PCR分析结果发现(图4),FtF3GT1基因表达表现出明显的组织差异和一定的生长发育时间差异。FtF3GT1基因在苦荞的各个组织中均有表达,且在茎中的表达量最高,约是根中表达量的70倍;在叶、花、不成熟种子和成熟种子中的表达量也高,但在根中表达量最低。
图4
图4
FtF3GT1基因的组织特异性表达
FtH3作为内参基因,每组数据代
Fig.4
Relative expression levels of FtF3GT1 gene in different tissues
FtH3 was used as an internal control. Error bars represent the standard deviation of triplicate runs for qRT-PCR. * indicate significant differences using t-test (P < 0.05), the same below
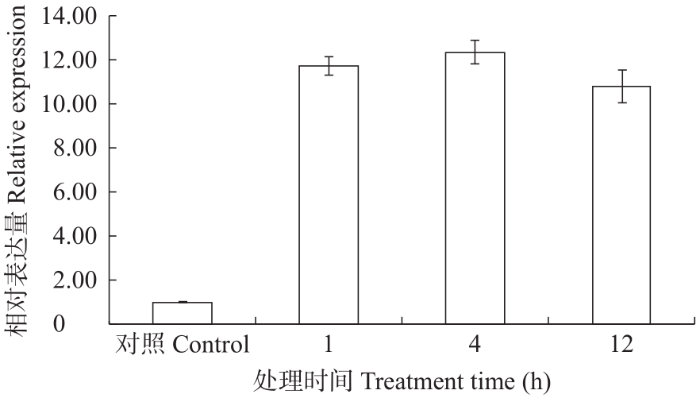
2.3.2 不同激素处理下FtF3GT1基因表达分析 对FtF3GT1基因的启动子进行分析,发现其启动子上有响应外源激素MeJA的作用元件,利用qRT-PCR技术分析了用50µmol/L MeJA处理7d的无菌苗,分析探索了FtF3GT1基因在MeJA胁迫处理下表达模式。结果(图5)表明,FtF3GT1基因的表达量与转录组数据总体上一致,随着MeJA处理时间的增加而逐渐增高,且在4h时达到最高,12h后有所下降。
图5
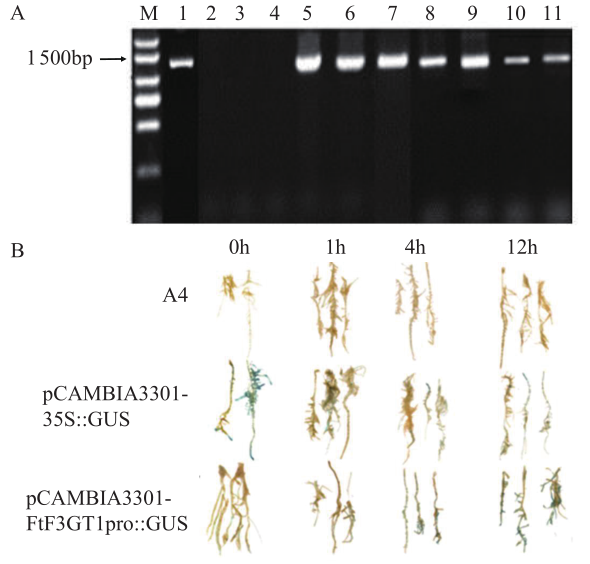
GUS染色结果(图6B)表明,pCAMBIA3301-35S::GUS毛状根染色最深,GUS活性最强,而pCAMBIA3301:FtF3GT1pro::GUS染色程度和GUS活性其次,但WT毛状根中没有发现有GUS活性。同时,随着MeJA处理时间的增长,pCAMBIA3301:FtF3GT1pro::GUS毛状根中GUS活性逐渐增强,表明获得的FtF3GT1基因启动子具有启动子的活性,能够启动GUS报告基因的表达,且其表达受MeJA的诱导。
图6
图6
转基因根系的鉴定及GUS染色
A:FtF3GT1pro::GUS转基因毛状根鉴定图;M:DL 2 000 DNA标记;1:pCAMBIA3301:FtF3GT1pro::GUS质粒;2~4:转A4根系DNA;5~7:转pCAMBIA3301-35S::GUS根系DNA;8~11:转FtF3GT1pro::GUS根系DNA;B:MeJA处理FtF3GT1pro::GUS毛状根的GUS染色情况
Fig.6
Identification of transgenic roots and GUS staining
A: Molecular identification of transgenic FtF3GT1pro::GUS hairy root lines. M: DL 2 000 DNA marker; 1: pCAMBIA3301:FtF3GT1pro::GUS plasmid; 2-4: transgenic lines DNA of A4 rhizogenes; 5-7: transgenic lines DNA of pCAMBIA3301-35S::GUS; 8-11: transgenic lines DNA of FtF3GT1pro::GUS; B: GUS staining of FtF3GT1pro::GUS hairy root treated with MeJA
2.4 毛状根的诱导及转基因根系的鉴定
图7
图7
苦荞毛状根的诱导过程和转基因根系的鉴定
A:苦荞毛状根的诱导过程;B:转基因根系的鉴定;M:DL 2 000 DNA标记;1:pCAMBIA1307-FtRT1质粒;2:天然的苦荞根系(WT);3:转pCAMBIA1307空载体根系;4~8:转pCAMBIA1307-FtRT1根系DNA检测
Fig.7
Induction process of hairy roots of tartary buckwheat and identification of transgenic root lines
A: Induction process of hairy roots of tartary buckwheat; B: Identification of transgenic root lines; M: DL 2 000 DNA Marker; 1: pCAMBIA1307-FtRT1 plasmid; 2: natural root of tartary buckwheat (WT); 3: pCAMBIA1307 transgenic root lines; 4-8: DNA detection of pCAMBIA1307-FtRT1 transgenic root lines
2.5 FtF3GT1转基因毛状根中总黄酮含量的变化
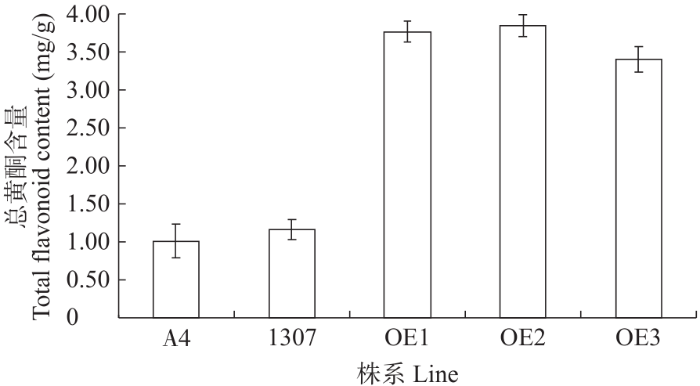
图8表明,3个过表达株系中总黄酮含量明显比对照组、转空载体毛状根中的总黄酮含量高3~4倍,表明FtF3GT1基因可能参与调控黄酮类物质的合成。
图8
图8
A4、pCAMBIA1307空载体和pCAMBIA1307-FtF3GT1过表达根系中总黄酮的测定
Fig.8
Determination of total flavonoids in A4, pCAMBIA1307 empty vector and pCAMBIA1307-FtF3GT1 in overexpression of root system
3 讨论
目前,已经有许多的研究表明GTs可以在植物次生代谢的合成中发挥重要作用。蓝花龙胆中的GtUGT7已被证实在植物体内具有有效的酶活性,能将一部分葡萄糖转移到花青素中3-糖苷[22];对影响翠蝶花花色形成的研究发现,鼠李糖基转移酶参与花青素修饰的功能表征,对花的颜色有很大的影响[23];在矮牵牛中还鉴定了一种仅在雄配子体中表达的黄酮醇3-O-半乳糖转移酶(F3GalTase),它能控制花粉特异性类糖基黄酮醇的形成[24]。根据类黄酮糖基化的区域选择性,将糖基转移酶分为3类:UF3GT、UF5GT和UF7GT[25]。基于UFGTs氨基酸序列的系统发育分析发现,FtF3GT1与FeF3G6″RhaT(FeUF7GT)具有极高的同源性。根据GT空间结构的不同,GTs又可以分为GT-A型折叠和GT-B型折叠,大多数的空间结构为GT-B型[26]。大多数的GT-B型糖基转移酶的N端同源性较低,C端的同源性较高,通常在C末端存在一个由44个氨基酸残基构成,被认为是糖基转移酶识别、结合供体分子位点的UGT保守序列(putative secondary plant glycosyltransferase,PSPG box),其中氨基酸序列HCGWNS高度保守[27]。多重序列比对分析表明,FtF3GT1是一个典型的GT-B型糖基转移酶。已有研究表明,FtF3GT1的同源蛋白FeUF7GT也是一个糖基转移酶,在甜荞黄酮合成途径中可以将槲皮素催化为芦丁[28],猜测FtF3GT1基因可能具有与FeUF7GT相同的生物学功能,这还需要后续的试验探究。
植物产生茉莉酸以调节各种植物过程,包括对伤害和非生物胁迫的反应,以及对昆虫和坏死性病原体的防御等[29]。茉莉酸及其循环前体和衍生物,统称为JAs,构成了一类已被发现可诱导编码酶的基因表达的氧化脂质,催化各种次生代谢物的形成[30]。有研究发现,JAs作为植物次生代谢产物合成途径最有效的诱导子,对植物的生长发育和防御机制具有重要的调控作用,如在转基因烟草植物表达拟南芥转录因子AtMYB12中发现,与非转基因植物相比,棉铃虫表现出更强的抗性,这可能是由于这些植物增强了芦丁的积累[31];Zhang等[32]利用转录组测序和重测序技术,分离并克隆了一组受JA诱导的参与调控芦丁合成的基因,证明MeJA参与芦丁的生物合成途径,它可以在蛋白或转录水平调控芦丁合成通路中的TFs或关键酶。JAs主要是通过与相关转录因子结合来共同调控一些次生代谢产物生物合成,其中一些转录因子可以调控多个生物合成途径相关基因的表达[29]。
本研究发现FtF3GT1基因对茉莉酸信号非常敏感,MeJA处理苦荞7d的无菌苗,FtF3GT1基因的表达量在4h时达到最高,比对照组上调了12倍左右,12h后略有下降的趋势。同时,转基因毛状根系中的总黄酮含量比野生型上调了4倍。表明其类黄酮含量高可能会增强自身对环境胁迫的适应性,对应的类黄酮含量也高。此外,随着MeJA处理时间的增长,转pCAMBIA3301:FtF3GT1pro::GUS毛状根中GUS活性也逐渐增强,证明MeJA影响了代谢途径中FtF3GT1基因的转录和翻译。Zhang等[32]的qRT-PCR分析显示,荞麦的转录因子FtMYB13、FtMYB14、FtMYB15和FtMYB16在不同组织中的转录积累模式不同;Zhang等[33]研究发现,FtMYB116也表现出组织表达特异性,主要在叶片中表达,在根中的表达量最低。此次研究发现FtF3GT1基因具有明显的器官特异性,在许多组织中都有表达,且FtF3GT1基因总mRNA在茎中积累最多,在根中积累最少。
4 结论
FtF3GT1基因在苦荞茎、叶、花和种子等组织中均有表达。过表达FtF3GT1基因,转基因根系中的总黄酮含量得到了显著提高。MeJA处理苦荞和pCAMBIA3301:FtF3GT1pro::GUS毛状根试验发现,FtF3GT1基因的表达量和GUS活性明显受MeJA诱导;且随着MeJA诱导时间的增长,转基因毛状根中的GUS活性增强。上述结果证明了FtF3GT1基因受MeJA诱导来调控类黄酮的生物合成。
参考文献
Chemopreventive effect of natural dietary compounds on xenobiotic-induced toxicity
Dietary flavonoids:Effects on xenobiotic and carcinogen metabolism
DOI:10.1016/j.tiv.2005.06.048
URL
PMID:16289744
[本文引用: 1]

Flavonoids are present in fruits, vegetables and beverages derived from plants (tea, red wine), and in many dietary supplements or herbal remedies including Ginkgo Biloba, Soy Isoflavones, and Milk Thistle. Flavonoids have been described as health-promoting, disease-preventing dietary supplements, and have activity as cancer preventive agents. Additionally, they are extremely safe and associated with low toxicity, making them excellent candidates for chemopreventive agents. The cancer protective effects of flavonoids have been attributed to a wide variety of mechanisms, including modulating enzyme activities resulting in the decreased carcinogenicity of xenobiotics. This review focuses on the flavonoid effects on cytochrome P450 (CYP) enzymes involved in the activation of procarcinogens and phase II enzymes, largely responsible for the detoxification of carcinogens. A number of naturally occurring flavonoids have been shown to modulate the CYP450 system, including the induction of specific CYP isozymes, and the activation or inhibition of these enzymes. Some flavonoids alter CYPs through binding to the aryl hydrocarbon receptor (AhR), a ligand-activated transcription factor, acting as either AhR agonists or antagonists. Inhibition of CYP enzymes, including CYP 1A1, 1A2, 2E1 and 3A4 by competitive or mechanism-based mechanisms also occurs. Flavones (chrysin, baicalein, and galangin), flavanones (naringenin) and isoflavones (genistein, biochanin A) inhibit the activity of aromatase (CYP19), thus decreasing estrogen biosynthesis and producing antiestrogenic effects, important in breast and prostate cancers. Activation of phase II detoxifying enzymes, such as UDP-glucuronyl transferase, glutathione S-transferase, and quinone reductase by flavonoids results in the detoxification of carcinogens and represents one mechanism of their anticarcinogenic effects. A number of flavonoids including fisetin, galangin, quercetin, kaempferol, and genistein represent potent non-competitive inhibitors of sulfotransferase 1A1 (or P-PST); this may represent an important mechanism for the chemoprevention of sulfation-induced carcinogenesis. Importantly, the effects of flavonoids on enzymes are generally dependent on the concentrations of flavonoids present, and the different flavonoids ingested. Due to the low oral bioavailability of many flavonoids, the concentrations achieved in vivo following dietary administration tend to be low, and may not reflect the concentrations tested under in vitro conditions; however, this may not be true following the ingestion of herbal preparations when much higher plasma concentrations may be obtained. Effects will also vary with the tissue distribution of enzymes, and with the species used in testing since differences between species in enzyme activities also can be substantial. Additionally, in humans, marked interindividual variability in drug-metabolizing enzymes occurs as a result of genetic and environmental factors. This variability in xenobiotic metabolizing enzymes and the effect of flavonoid ingestion on enzyme expression and activity can contribute to the varying susceptibility different individuals have to diseases such as cancer. As well, flavonoids may also interact with chemotherapeutic drugs used in cancer treatment through the induction or inhibition of their metabolism.
Antimicrobial activity of flavonoids
DOI:10.1016/j.ijantimicag.2005.09.002
URL
PMID:16323269
[本文引用: 1]

Flavonoids are ubiquitous in photosynthesising cells and are commonly found in fruit, vegetables, nuts, seeds, stems, flowers, tea, wine, propolis and honey. For centuries, preparations containing these compounds as the principal physiologically active constituents have been used to treat human diseases. Increasingly, this class of natural products is becoming the subject of anti-infective research, and many groups have isolated and identified the structures of flavonoids possessing antifungal, antiviral and antibacterial activity. Moreover, several groups have demonstrated synergy between active flavonoids as well as between flavonoids and existing chemotherapeutics. Reports of activity in the field of antibacterial flavonoid research are widely conflicting, probably owing to inter- and intra-assay variation in susceptibility testing. However, several high-quality investigations have examined the relationship between flavonoid structure and antibacterial activity and these are in close agreement. In addition, numerous research groups have sought to elucidate the antibacterial mechanisms of action of selected flavonoids. The activity of quercetin, for example, has been at least partially attributed to inhibition of DNA gyrase. It has also been proposed that sophoraflavone G and (-)-epigallocatechin gallate inhibit cytoplasmic membrane function, and that licochalcones A and C inhibit energy metabolism. Other flavonoids whose mechanisms of action have been investigated include robinetin, myricetin, apigenin, rutin, galangin, 2,4,2'-trihydroxy-5'-methylchalcone and lonchocarpol A. These compounds represent novel leads, and future studies may allow the development of a pharmacologically acceptable antimicrobial agent or class of agents.
Recent advances in understanding the antibacterial properties of flavonoids
DOI:10.1016/j.ijantimicag.2011.02.014
URL
[本文引用: 1]

Antibiotic resistance is a major global problem and there is a pressing need to develop new therapeutic agents. Flavonoids are a family of plant-derived compounds with potentially exploitable activities, including direct antibacterial activity, synergism with antibiotics, and suppression of bacterial virulence. In this review, recent advances towards understanding these properties are described. Information is presented on the ten most potently antibacterial flavonoids as well as the five most synergistic flavonoid-antibiotic combinations tested in the last 6 years (identified from PubMed and Science-Direct). Top of these respective lists are panduratin A, with minimum inhibitory concentrations (MICs) of 0.06-2.0 mu g/mL against Staphylococcus aureus, and epicatechin gallate, which reduces oxacillin MICs as much as 512-fold. Research seeking to improve such activity and understand structure-activity relationships is discussed. Proposed mechanisms of action are also discussed. In addition to direct and synergistic activities, flavonoids inhibit a number of bacterial virulence factors, including quorum-sensing signal receptors, enzymes and toxins. Evidence of these molecular effects at the cellular level include in vitro inhibition of biofilm formation, inhibition of bacterial attachment to host ligands, and neutralisation of toxicity towards cultured human cells. In vivo evidence of disruption of bacterial pathogenesis includes demonstrated efficacy against Helicobacter pylori infection and S. aureus alpha-toxin intoxication. (c) 2011 Elsevier B. V. and the International Society of Chemotherapy.
Flavonoids:new roles for old molecules
DOI:10.1111/j.1744-7909.2010.00905.x
URL
[本文引用: 1]

Arabidopsis thaliana and Medicago truncatula are enabling the intricacies of the physiology of these compounds to be deduced. In the present review, we cover recent advances in flavonoid research, discuss deficiencies in our understanding of the physiological processes, and suggest approaches to identify the cellular targets of flavonoids.Buer CS, Imin N, Djordjevic MA (2010) Flavonoids: new roles for old molecules. J. Integr. Plant Biol. 52(1), 98–111.]]>
Rutin,quercetin,and free amino acid analysis in buckwheat (Fagopyrum) seeds from different locations
DOI:10.4238/2015.December.29.11
URL
PMID:26782554
[本文引用: 1]

In this study, five common buckwheats and nine tartary buckwheats grown at different locations were analyzed for the contents of rutin, quercetin, and amino acids by high-performance liquid chromatography and spectrophotometry. The rutin content was higher than quercetin in buckwheat seeds. Rutin content was in the range from 0.05 (0.05 g per 100 g dry seeds) to 1.35% of buckwheat seeds. Quercetin content varied from 0.01 to 0.17% and in some common buckwheats it was even difficult to detect. Comparatively, tartary buckwheat seeds contained more rutin and quercetin than common buckwheat seeds. Meanwhile, the bran has higher rutin content than the farina in tartary buckwheat seeds, with a respective content of 0.45 to 1.19% and 0.14 to 0.67%. It was found that amino acid contents were around 1.79 to 12.65% (farina) and 5.74 to 7.89% (bran) in common buckwheats, and 1.73 to 5.63% (farina) and 2.64 to 16.78% (bran) in tartary buckwheat seeds. The highest total rutin content was found to be 1.35% in tartary buckwheat seeds from Sichuan, China. The highest total amounts of amino acid were detected to be 20.13% in tartary buckwheat seeds from Changzhi, Shanxi Province (China). Our results suggested that food products made of whole-buckwheat flour are healthier than those made of fine white flour.
FtSAD2 and FtJAZ1 regulate activity of the FtMYB11 transcription repressor of the phenylpropanoid pathway in Fagopyrum tataricum
DOI:10.1111/nph.14692
URL
PMID:28722263
[本文引用: 1]

Little is known about the molecular mechanism of the R2R3-MYB transcriptional repressors involved in plant phenylpropanoid metabolism. Here, we describe one R2R3-type MYB repressor, FtMYB11 from Fagopyrum tataricum. It contains the SID-like motif GGDFNFDL and it is regulated by both the importin protein 'Sensitive to ABA and Drought 2' (SAD2) and the jasmonates signalling cascade repressor JAZ protein. Yeast two hybrid and bimolecular fluorescence complementation assays demonstrated that FtMYB11 interacts with SAD2 and FtJAZ1. Protoplast transactivation assays demonstrated that FtMYB11 acts synergistically with FtSAD2 or FtJAZ1 and directly represses its target genes via the MYB-core element AATAGTT. Changing the Asp122 residue to Asn in the SID-like motif results in cytoplasmic localization of FtMYB11 because of loss of interaction with SAD2, while changing the Asp126 residue to Asn results in the loss of interaction with FtJAZ1. Overexpression of FtMYB11or FtMYB11(D126N) in F. tataricum hairy roots resulted in reduced accumulation of rutin, while overexpression of FtMYB11(D122N) in hairy roots did not lead to such a change. The results indicate that FtMYB11 acts as a regulator via interacting with FtSAD2 or FtJAZ1 to repress phenylpropanoid biosynthesis, and this repression depends on two conserved Asp residues of its SID-like motif.
Jasmonate-responsive transcription factors regulating plant secondary metabolism
DOI:10.1016/j.biotechadv.2016.02.004
URL
PMID:26876016
[本文引用: 1]

Plants produce a large variety of secondary metabolites including alkaloids, glucosinolates, terpenoids and phenylpropanoids. These compounds play key roles in plant-environment interactions and many of them have pharmacological activity in humans. Jasmonates (JAs) are plant hormones which induce biosynthesis of many secondary metabolites. JAs-responsive transcription factors (TFs) that regulate the JAs-induced accumulation of secondary metabolites belong to different families including AP2/ERF, bHLH, MYB and WRKY. Here, we give an overview of the types and functions of TFs that have been identified in JAs-induced secondary metabolite biosynthesis, and highlight their similarities and differences in regulating various biosynthetic pathways. We review major recent developments regarding JAs-responsive TFs mediating secondary metabolite biosynthesis, and provide suggestions for further studies.
MdMYB9 and MdMYB11 are involved in the regulation of the JA-induced biosynthesis of anthocyanin and proanthocyanidin in apples
DOI:10.1093/pcp/pcu205
URL
PMID:25527830
[本文引用: 1]

Anthocyanin and proanthocyanidin (PA) are important secondary metabolites and beneficial to human health. Their biosynthesis is induced by jasmonate (JA) treatment and regulated by MYB transcription factors (TFs). However, which and how MYB TFs regulate this process is largely unknown in apple. In this study, MdMYB9 and MdMYB11 which were induced by methyl jasmonate (MeJA) were functionally characterized. Overexpression of MdMYB9 or MdMYB11 promoted not only anthocyanin but also PA accumulation in apple calluses, and the accumulation was further enhanced by MeJA. Subsequently, yeast two-hybrid, pull-down and bimolecular fluorescence complementation assays showed that both MYB proteins interact with MdbHLH3. Moreover, Jasmonate ZIM-domain (MdJAZ) proteins interact with MdbHLH3. Furthermore, chromatin immunoprecipitation-quantitative PCR and yeast one-hybrid assays demonstrated that both MdMYB9 and MdMYB11 bind to the promoters of ANS, ANR and LAR, whereas MdbHLH3 is recruited to the promoters of MdMYB9 and MdMYB11 and regulates their transcription. In addition, transient expression assays indicated that overexpression of MdJAZ2 inhibits the recruitment of MdbHLH3 to the promoters of MdMYB9 and MdMYB11. Our findings provide new insight into the mechanism of how MeJA regulates anthocyanin and PA accumulation in apple.
Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings
DOI:10.1111/j.1365-313X.2007.03373.x
URL
PMID:18036197
[本文引用: 1]

In all higher plants studied to date, the anthocyanin pigment pathway is regulated by a suite of transcription factors that include Myb, bHLH and WD-repeat proteins. However, in Arabidopsis thaliana, the Myb regulators remain to be conclusively identified, and little is known about anthocyanin pathway regulation by TTG1-dependent transcriptional complexes. Previous overexpression of the PAP1 Myb suggested that genes from the entire phenylpropanoid pathway are targets of regulation by Myb/bHLH/WD-repeat complexes in Arabidopsis, in contrast to other plants. Here we demonstrate that overexpression of Myb113 or Myb114 results in substantial increases in pigment production similar to those previously seen as a result of over-expression of PAP1, and pigment production in these overexpressors remains TTG1- and bHLH-dependent. Also, plants harboring an RNAi construct targeting PAP1 and three Myb candidates (PAP2, Myb113 and Myb114) showed downregulated Myb gene expression and obvious anthocyanin deficiencies. Correlated with these anthocyanin deficiencies is downregulation of the same late anthocyanin structural genes that are downregulated in ttg1 and bHLH anthocyanin mutants. Expression studies using GL3:GR and TTG1:GR fusions revealed direct regulation of the late biosynthetic genes only. Functional diversification between GL3 and EGL3 with regard to activation of gene targets was revealed by GL3:GR studies in single and double bHLH mutant seedlings. Expression profiles for Myb and bHLH regulators are also presented in the context of pigment production in young seedlings.
Filamentous flower is a direct target of JAZ3 and modulates responses to jasmonate
DOI:10.1105/tpc.15.00220
URL
PMID:26530088
[本文引用: 1]

The plant hormone jasmonate (JA) plays an important role in regulating growth, development, and immunity. Activation of the JA-signaling pathway is based on the hormone-triggered ubiquitination and removal of transcriptional repressors (JASMONATE-ZIM DOMAIN [JAZ] proteins) by an SCF receptor complex (SCF(COI1)/JAZ). This removal allows the rapid activation of transcription factors (TFs) triggering a multitude of downstream responses. Identification of TFs bound by the JAZ proteins is essential to better understand how the JA-signaling pathway modulates and integrates different responses. In this study, we found that the JAZ3 repressor physically interacts with the YABBY (YAB) family transcription factor FILAMENTOUS FLOWER (FIL)/YAB1. In Arabidopsis thaliana, FIL regulates developmental processes such as axial patterning and growth of lateral organs, shoot apical meristem activity, and inflorescence phyllotaxy. Phenotypic analysis of JA-regulated responses in loss- and gain-of-function FIL lines suggested that YABs function as transcriptional activators of JA-triggered responses. Moreover, we show that MYB75, a component of the WD-repeat/bHLH/MYB complex regulating anthocyanin production, is a direct transcriptional target of FIL. We propose that JAZ3 interacts with YABs to attenuate their transcriptional function. Upon perception of JA signal, degradation of JAZ3 by the SCF(COI1) complex releases YABs to activate a subset of JA-regulated genes in leaves leading to anthocyanin accumulation, chlorophyll loss, and reduced bacterial defense.
A radish basic helix-loop-helix transcription factor,RsTT8 acts a positive regulator for anthocyanin biosynthesis
DOI:10.3389/fpls.2017.01917
URL
PMID:29167678
[本文引用: 1]

The MYB-bHLH-WDR (MBW) complex activates anthocyanin biosynthesis through the transcriptional regulation. RsMYB1 has been identified as a key player in anthocyanin biosynthesis in red radish (Raphanus sativus L.), but its partner bHLH transcription factor (TF) remains to be determined. In this study, we isolated a bHLH TF gene from red radish. Phylogenetic analysis indicated that this gene belongs to the TT8 clade of the IIIF subgroup of bHLH TFs, and we thus designated this gene RsTT8. Subcellular localization analysis showed that RsTT8-sGFP was localized to the nuclei of Arabidopsis thaliana protoplasts harboring the RsTT8-sGFP construct. We evaluated anthocyanin biosynthesis and RsTT8 expression levels in three radish varieties (N, C, and D) that display different red phenotypes in the leaves, root flesh, and root skins. The root flesh of the C variety and the leaves and skins of the D variety exhibit intense red pigmentation; in these tissues, RsTT8 expression showed totally positive association with the expression of RsMYB1 TF and of five of eight tested anthocyanin biosynthesis genes (i.e., RsCHS, RsCHI, RsF3H, RsDFR, and RsANS). Heterologous co-expression of both RsTT8 and RsMYB1 in tobacco leaves dramatically increased the expression of endogenous anthocyanin biosynthesis genes and anthocyanin accumulation. Furthermore, a yeast two-hybrid assay showed that RsTT8 interacts with RsMYB1 at the MYB-interacting region (MIR), and a transient transactivation assay indicated that RsTT8 activates the RsCHS and RsDFR promoters when co-expressed with RsMYB1. Complementation of the Arabidopsis tt8-1 mutant, which lacks red pigmentation in the leaves and seeds, with RsTT8 restored red pigmentation, and resulted in high anthocyanin and proanthocyanidin contents in the leaves and seeds, respectively. Together, these results show that RsTT8 functions as a regulatory partner with RsMYB1 during anthocyanin biosynthesis.
The evolution of phenylpropanoid metabolism in the green lineage
DOI:10.3109/10409238.2012.758083
URL
[本文引用: 1]

Phenolic secondary metabolites are only produced by plants wherein they play important roles in both biotic and abiotic defense in seed plants as well as being potentially important bioactive compounds with both nutritional and medicinal benefits reported for animals and humans as a consequence of their potent antioxidant activity. During the long evolutionary period in which plants have adapted to the environmental niches in which they exist (and especially during the evolution of land plants from their aquatic algal ancestors), several strategies such as gene duplication and convergent evolution have contributed to the evolution of this pathway. In this respect, diversity and redundancy of several key genes of phenolic secondary metabolism such as polyketide synthases, cytochrome P450s, Fe2+/2-oxoglutarate-dependent dioxygenases and UDP-glycosyltransferases have played an essential role. Recent technical developments allowing affordable whole genome sequencing as well as a better inventory of species-by-species chemical diversity have resulted in a dramatic increase in the number of tools we have to assess how these pathways evolved. In parallel, reverse genetics combined with detailed molecular phenotyping is allowing us to elucidate the functional importance of individual genes and metabolites and by this means to provide further mechanistic insight into their biological roles. In this review, phenolic metabolite-related gene sequences (for a total of 65 gene families including shikimate biosynthetic genes) are compared across 23 independent species, and the phenolic metabolic complement of various plant species are compared with one another, in attempt to better understand the evolution of diversity in this crucial pathway.
Biosynthesis of anthocyanins and their regulation in colored grapes
DOI:10.3390/molecules15129057
URL
PMID:21150825
[本文引用: 1]

Anthocyanins, synthesized via the flavonoid pathway, are a class of crucial phenolic compounds which are fundamentally responsible for the red color of grapes and wines. As the most important natural colorants in grapes and their products, anthocyanins are also widely studied for their numerous beneficial effects on human health. In recent years, the biosynthetic pathway of anthocyanins in grapes has been thoroughly investigated. Their intracellular transportation and accumulation have also been further clarified. Additionally, the genetic mechanism regulating their biosynthesis and the phytohormone influences on them are better understood. Furthermore, due to their importance in the quality of wine grapes, the effects of the environmental factors and viticulture practices on anthocyanin accumulation are being investigated increasingly. The present paper summarizes both the basic information and the most recent advances in the study of the anthocyanin biosynthesis in red grapes, emphasizing their gene structure, the transcriptional factors and the diverse exterior regulation factors.
Genetic and metabolic engineering of isoflavonoid biosynthesis
Purification of RNA using trizol
Transcriptional response of the catharanthine biosynthesis pathway to methyl jasmonate/nitric oxide elicitation in Catharanthus roseus hairy root culture
DOI:10.1007/s00253-010-2822-x
URL
[本文引用: 1]

CrORCA3 (octadecanoid-responsive Catharanthus AP2/ERF domain), methyl jasmonate (MeJA), and sodium nitroprusside (SNP) on the differentiated tissue of Catharanthus roseus hairy roots. The changes in catharanthine concentration and in the levels of mRNA transcripts of pathway genes and regulators were tracked for 192 h. ORCA3 overexpression led to a slight decrease of the accumulation of catharanthine, while MeJA treatment caused a large increase in the levels of transcripts of pathway genes and the catharanthine concentration. SNP treatment alone or SNP in combination with MeJA treatment caused a dramatic decrease of the cathanranthine concentration, while at the same time the levels of transcripts of zinc finger-binding proteins genes (ZCTs) increased. The latter treatment also caused a decrease of the levels of transcripts of type-I protein prenyltransferase gene (PGGT-I). This response of transcriptional repressors and pathway genes may explain the antagonistic effects of NO and MeJA on catharanthine biosynthesis in C. roseus hairy roots.]]>
Cloning and characterization of the UDP-glucose:anthocyanin 5-O-glucosyltransferase gene from blue-flowered gentian
DOI:10.1093/jxb/ern031
URL
PMID:18375606
[本文引用: 1]

Blue-flowered gentian (Gentiana triflora) is known to accumulate gentiodelphin, a unique polyacylated delphinidin-type anthocyanin, in the petals. Almost all of the structural genes involved in gentiodelphin biosynthesis have been isolated, but an important gene encoding UDP-glucose:anthocyanin 5-O-glucosyltransferase (5GT) remained to be identified. In this study, an attempt was made to isolate and characterize gentian 5GT, which is responsible for glucosylation of anthocyanidin 3-glucoside. A PCR-based cloning strategy identified seven 5GT candidates from gentian flowers. Among them, the deduced amino acid sequence of the 5GT gene from gentian petal cDNA, designated Gt5GT7, exhibited 36.0-41.7% identities with those of 5GTs from other plant species, and phylogenic analysis also suggested that Gt5GT7 belongs to the 5GT subfamily. The expression analysis showed that Gt5GT7 transcripts were detected predominantly in petals and weakly in filaments but not in leaves, stems, and other floral organs. In addition, increased levels of Gt5GT7 transcripts in petals coincided with flower development, a pattern identical to that of 5GT enzymatic activity as determined by in vitro assay using petal crude proteins. The substrate specificity of Gt5GT7 was analysed in vitro using the recombinant enzyme produced by Escherichia coli. Gt5GT7 could transfer a glucosyl moiety to anthocyanidin 3-glycosides but not to other flavonoid compounds. Delphinidin 3-glucoside, the precursor of gentiodelphin, was the best substrate among several anthocyanidin 3-glycosides tested. Heterologous expression of Gt5GT7 in tobacco plants led to additional accumulation of cyanidin 3-rutinoside-5-glucoside, confirming that Gt5GT7 has a valid enzymatic activity in planta.
Functional characterization of UDP-rhamnose-dependent rhamnosyltransferase involved in anthocyanin modification,a key enzyme determining blue coloration in Lobelia erinus
DOI:10.1111/tpj.13387
URL
PMID:27696560
[本文引用: 1]

Because structural modifications of flavonoids are closely related to their properties, such as stability, solubility, flavor and coloration, characterizing the enzymes that catalyze the modification reactions can be useful for engineering agriculturally beneficial traits of flavonoids. In this work, we examined the enzymes involved in the modification pathway of highly glycosylated and acylated anthocyanins that accumulate in Lobelia erinus. Cultivar Aqua Blue (AB) of L. erinus is blue-flowered and accumulates delphinidin 3-O-p-coumaroylrutinoside-5-O-malonylglucoside-3'5'-O-dihydroxycinnamoylglucoside (lobelinins) in its petals. Cultivar Aqua Lavender (AL) is mauve-flowered, and LC-MS analyses showed that AL accumulated delphinidin 3-O-glucoside (Dp3G), which was not further modified toward lobelinins. A crude protein assay showed that modification processes of lobelinin were carried out in a specific order, and there was no difference between AB and AL in modification reactions after rhamnosylation of Dp3G, indicating that the lack of highly modified anthocyanins in AL resulted from a single mutation of rhamnosyltransferase catalyzing the rhamnosylation of Dp3G. We cloned rhamnosyltransferase genes (RTs) from AB and confirmed their UDP-rhamnose-dependent rhamnosyltransferase activities on Dp3G using recombinant proteins. In contrast, the RT gene in AL had a 5-bp nucleotide deletion, resulting in a truncated polypeptide without the plant secondary product glycosyltransferase box. In a complementation test, AL that was transformed with the RT gene from AB produced blue flowers. These results suggest that rhamnosylation is an essential process for lobelinin synthesis, and thus the expression of RT has a great impact on the flower color and is necessary for the blue color of Lobelia flowers.
Purification,cloning,and heterologous expression of a catalytically efficient flavonol 3-O-galactosyltransferase expressed in the male gametophyte of Petunia hybrida
DOI:10.1074/jbc.274.48.34011
URL
PMID:10567367
[本文引用: 1]

Flavonols are plant-specific molecules that are required for pollen germination in maize and petunia. They exist in planta as both the aglycone and glycosyl conjugates. We identified a flavonol 3-O-galactosyltransferase (F3GalTase) that is expressed exclusively in the male gametophyte and controls the formation of a pollen-specific class of glycosylated flavonols. Thus an essential step to understanding flavonol-induced germination is the characterization of F3GalTase. Amino acid sequences of three peptide fragments of F3GalTase purified from petunia pollen were used to isolate a full-length cDNA clone. RNA gel blot analysis and enzyme assays confirmed that F3GalTase expression is restricted to pollen. Heterologous expression of the F3GalTase cDNA in Escherichia coli yielded active recombinant enzyme (rF3GalTase) which had the identical substrate specificity as the native enzyme. Unlike the relatively nonspecific substrate usage of flavonoid glycosyltransferases from sporophytic tissues, F3GalTase uses only UDP-galactose and flavonols to catalyze the formation of flavonol 3-O-galactosides. Kinetic analysis showed that the k(cat)/K(m) values of rF3GalTase, using kaempferol and quercetin as substrates, approaches that of a catalytically perfect enzyme. rF3GalTase catalyzes the reverse reaction, generation of flavonols from UDP and flavonol 3-O-galactosides, almost as efficiently as the forward reaction. The biochemical characteristics of F3GalTase are discussed in the context of a role in flavonol-induced pollen germination.
Functional characterization of a glucosyltransferase gene,LcUFGT1,involved in the formation of cyanidin glucoside in the pericarp of Litchi chinensis
DOI:10.1111/ppl.12391
URL
PMID:26419221
[本文引用: 1]

Anthocyanins generate the red color in the pericarp of Litchi chinensis. UDP-glucose: flavonoid 3-O-glycosyltransferase (UFGT, EC. 2.4.1.91) stabilizes anthocyanidin by attaching sugar moieties to the anthocyanin aglycone. In this study, the function of an UFGT gene involved in the biosynthesis of anthocyanin was verified through heterologous expression and virus-induced gene silencing assays. A strong positive correlation between UFGT activity and anthocyanin accumulation capacity was observed in the pericarp of 15 cultivars. Four putative flavonoid 3-O-glycosyltransferase-like genes, designated as LcUFGT1 to LcUFGT4, were identified in the pericarp of litchi. Among the four UFGT gene members, only LcUFGT1 can use cyanidin as its substrate. The expression of LcUFGT1 was parallel with developmental anthocyanin accumulation, and the heterologously expressed protein of LcUFGT1 displayed catalytic activities in the formation of anthocyanin. The LcUFGT1 over-expression tobacco had darker petals and pigmented filaments and calyxes resulting from higher anthocyanin accumulations compared with non-transformed tobacco. In the pericarp with LcUFGT1 suppressed by virus-induced gene silencing, pigmentation was retarded, which was well correlated with the reduced-LcUFGT1 transcriptional activity. These results suggested that the glycosylation-related gene LcUFGT1 plays a critical role in red color formation in the pericarp of litchi.
A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities
Identification and characterization of a rhamnosyltransferase involved in rutin biosynthesis in Fagopyrum esculentum (common buckwheat)
DOI:10.1080/09168451.2018.1491286
URL
PMID:29972345
[本文引用: 1]

Rutin, a 3-rutinosyl quercetin, is a representative flavonoid distributed in many plant species, and is highlighted for its therapeutic potential. In this study, we purified uridine diphosphate-rhamnose: quercetin 3-O-glucoside 6''-O-rhamnosyltransferase and isolated the corresponding cDNA (FeF3G6''RhaT) from seedlings of common buckwheat (Fagopyrum esculentum). The recombinant FeF3G6''RhaT enzyme expressed in Escherichia coli exhibited 6''-O-rhamnosylation activity against flavonol 3-O-glucoside and flavonol 3-O-galactoside as substrates, but showed only faint activity against flavonoid 7-O-glucosides. Tobacco cells expressing FeF3G6''RhaT converted the administered quercetin into rutin, suggesting that FeF3G6''RhaT can function as a rhamnosyltransferase in planta. Quantitative PCR analysis on several organs of common buckwheat revealed that accumulation of FeF3G6''RhaT began during the early developmental stages of rutin-accumulating organs, such as flowers, leaves, and cotyledons. These results suggest that FeF3G6''RhaT is involved in rutin biosynthesis in common buckwheat.
Jasmonate signalling:a copycat of auxin signalling?
DOI:10.1111/pce.12121
URL
PMID:23611666
[本文引用: 2]

Plant hormones regulate almost all aspects of plant growth and development. The past decade has provided breakthrough discoveries in phytohormone sensing and signal transduction, and highlighted the striking mechanistic similarities between the auxin and jasmonate (JA) signalling pathways. Perception of auxin and JA involves the formation of co-receptor complexes in which hormone-specific E3-ubiquitin ligases of the SKP1-Cullin-F-box protein (SCF) type interact with specific repressor proteins. Across the plant kingdom, the Aux/IAA and the JASMONATE-ZIM DOMAIN (JAZ) proteins correspond to the auxin- and JA-specific repressors, respectively. In the absence of the hormones, these repressors form a complex with transcription factors (TFs) specific for both pathways. They also recruit several proteins, among which the general co-repressor TOPLESS, and thereby prevent the TFs from activating gene expression. The hormone-mediated interaction between the SCF and the repressors targets the latter for 26S proteasome-mediated degradation, which, in turn, releases the TFs to allow modulating hormone-dependent gene expression. In this review, we describe the similarities and differences in the auxin and JA signalling cascades with respect to the protein families and the protein domains involved in the formation of the pathway-specific complexes.
ORCAnization of jasmonate-responsive gene expression in alkaloid metabolism
DOI:10.1016/s1360-1385(01)01924-0
URL
PMID:11335174
[本文引用: 1]

Jasmonic acid is an important plant stress signalling molecule. It induces the biosynthesis of defence proteins and protective secondary metabolites. In alkaloid metabolism, jasmonate acts by coordinate activation of the expression of multiple biosynthesis genes. In terpenoid indole alkaloid metabolism and primary precursor pathways, jasmonate induces gene expression and metabolism via ORCAs, which are members of the AP2/ERF-domain family of plant transcription factors. Other jasmonate-regulated (secondary) metabolic pathways might also be controlled by ORCA-like AP2/ERF-domain transcription factors. If so, such regulators could be used to improve plant fitness or metabolite productivity of plants or cell cultures.
Modulation of transcriptome and metabolome of tobacco by Arabidopsis transcription factor,AtMYB12,leads to insect resistance
DOI:10.1104/pp.109.150979
URL
PMID:20190095
[本文引用: 1]

Flavonoids synthesized by the phenylpropanoid pathway participate in myriad physiological and biochemical processes in plants. Due to the diversity of secondary transformations and the complexity of the regulation of branched pathways, single gene strategies have not been very successful in enhancing the accumulation of targeted molecules. We have expressed an Arabidopsis (Arabidopsis thaliana) transcription factor, AtMYB12, in tobacco (Nicotiana tabacum), which resulted in enhanced expression of genes involved in the phenylpropanoid pathway, leading to severalfold higher accumulation of flavonols. Global gene expression and limited metabolite profiling of leaves in the transgenic lines of tobacco revealed that AtMYB12 regulated a number of pathways, leading to flux availability for the phenylpropanoid pathway in general and flavonol biosynthesis in particular. The tobacco transgenic lines developed resistance against the insect pests Spodoptera litura and Helicoverpa armigera due to enhanced accumulation of rutin. Suppression of flavonol biosynthesis by artificial microRNA reversed insect resistance of the AtMYB12-expressing tobacco plants. Our study suggests that AtMYB12 can be strategically used for developing safer insect pest-resistant transgenic plants.
Jasmonate-responsive MYB factors spatially repress rutin biosynthesis in Fagopyrum tataricum
DOI:10.1093/jxb/ery032
URL
PMID:29394372
[本文引用: 2]

Jasmonates are plant hormones that induce the accumulation of many secondary metabolites, such as rutin in buckwheat, via regulation of jasmonate-responsive transcription factors. Here, we report on the identification of a clade of jasmonate-responsive subgroup 4 MYB transcription factors, FtMYB13, FtMYB14, FtMYB15, and FtMYB16, which directly repress rutin biosynthesis in Fagopyrum tataricum. Immunoblot analysis showed that FtMYB13, FtMYB14, and FtMYB15 could be degraded via the 26S proteasome in the COI1-dependent jasmonate signaling pathway, and that this degradation is due to the SID motif in their C-terminus. Yeast two-hybrid and bimolecular fluorescence complementation assays revealed that FtMYB13, FtMYB14, and FtMYB15 interact with the importin protein Sensitive to ABA and Drought 2 (FtSAD2) in stem and inflorescence. Furthermore, the key repressor of jasmonate signaling FtJAZ1 specifically interacts with FtMYB13. Point mutation analysis showed that the conserved Asp residue of the SID domain contributes to mediating protein-protein interaction. Protoplast transient activation assays demonstrated that FtMYB13, FtMYB14, and FtMYB15 directly repress phenylalanine ammonia lyase (FtPAL) gene expression, and FtSAD2 and FtJAZ1 significantly promote the repressing activity of FtMYBs. These findings may ultimately be promising for further engineering of plant secondary metabolism.
The light-induced transcription factor FtMYB116 promotes accumulation of rutin in Fagopyrum tataricum
DOI:10.1111/pce.13470
URL
PMID:30375656
[本文引用: 1]

Tartary buckwheat (Fagopyrum tataricum) not only provides a supplement to primary grain crops in China but also has high medicinal value, by virtue of its rich content of flavonoids possessing antioxidant, anti-inflammatory, and anticancer properties. Light is an important environmental factor that can regulate the synthesis of plant secondary metabolites. In this study, we treated tartary buckwheat seedlings with different wavelengths of light and found that red and blue light could increase the content of flavonoids and the expression of genes involved in flavonoid synthesis pathways. Through coexpression analysis, we identified a new MYB transcription factor (FtMYB116) that can be induced by red and blue light. Yeast one-hybrid assays and an electrophoretic mobility shift assay showed that FtMYB116 binds directly to the promoter region of flavonoid-3'-hydroxylase (F3'H), and a transient luciferase activity assay indicated that FtMYB116 can induce F3'H expression. After transforming FtMYB116 into the hairy roots of tartary buckwheat, we observed significant increases in the content of rutin and quercetin. Collectively, our results indicate that red and blue light promote an increase in flavonoid content in tartary buckwheat seedlings; we also identified a new MYB transcription factor, FtMYB116, that promotes the accumulation of rutin via direct activation of F3'H expression.