作物杂志,2025, 第4期: 29–40 doi: 10.16035/j.issn.1001-7283.2025.04.004
基于SSR技术的马铃薯种质资源遗传多样性分析
王丹1( ), 张志成2,3(
), 张志成2,3( ), 韩海霞1, 韩娜1, 王春勇4, 王正1, 潘孟洋1, 李强5, 王懿茜2, 刘宇飞2, 张丹2
), 韩海霞1, 韩娜1, 王春勇4, 王正1, 潘孟洋1, 李强5, 王懿茜2, 刘宇飞2, 张丹2
- 1集宁师范学院生命科学与技术学院,012000,内蒙古乌兰察布
2乌兰察布市农林科学研究所,012000,内蒙古乌兰察布
3内蒙古农业大学农学院,010000,内蒙古呼和浩特
4唐山市农业科学研究院,063000,河北唐山
5乌兰察布市农牧业综合行政执法支队,012000,内蒙古乌兰察布
Genetic Diversity Analysis of Potato Germplasm Resources Based on SSR Technology
Wang Dan1( ), Zhang Zhicheng2,3(
), Zhang Zhicheng2,3( ), Han Haixia1, Han Na1, Wang Chunyong4, Wang Zheng1, Pan Mengyang1, Li Qiang5, Wang Yiqian2, Liu Yufei2, Zhang Dan2
), Han Haixia1, Han Na1, Wang Chunyong4, Wang Zheng1, Pan Mengyang1, Li Qiang5, Wang Yiqian2, Liu Yufei2, Zhang Dan2
- 1Jining Normal University, Ulanqab 012000, Inner Mongolia, China
2Ulanqab Institute of Agricultural and Forestry Sciences, Ulanqab 012000, Inner Mongolia, China
3Inner Mongolia Agricultural University, Hohhot 010000, Inner Mongolia, China
4Tangshan Academy of Agricultural Sciences, Tangshan 063000, Hebei, China
5Ulanqab Agricultural and Animal Husbandry Comprehensive Administrative Law Enforcement Detachment, Ulanqab 012000, Inner Mongolia, China
摘要:
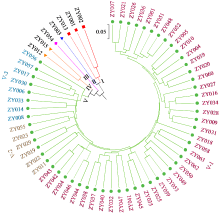
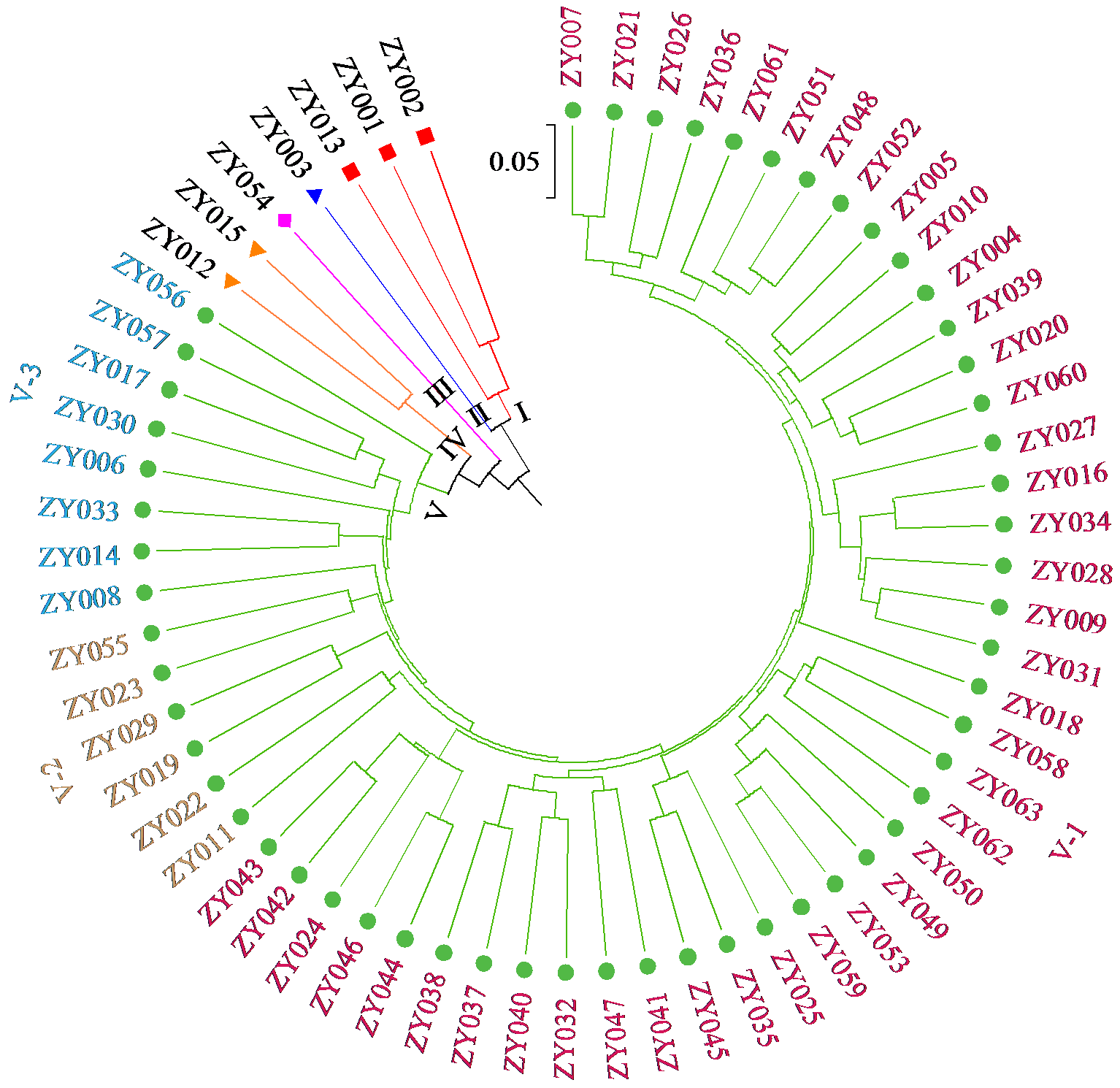
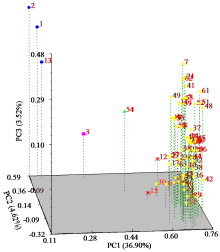
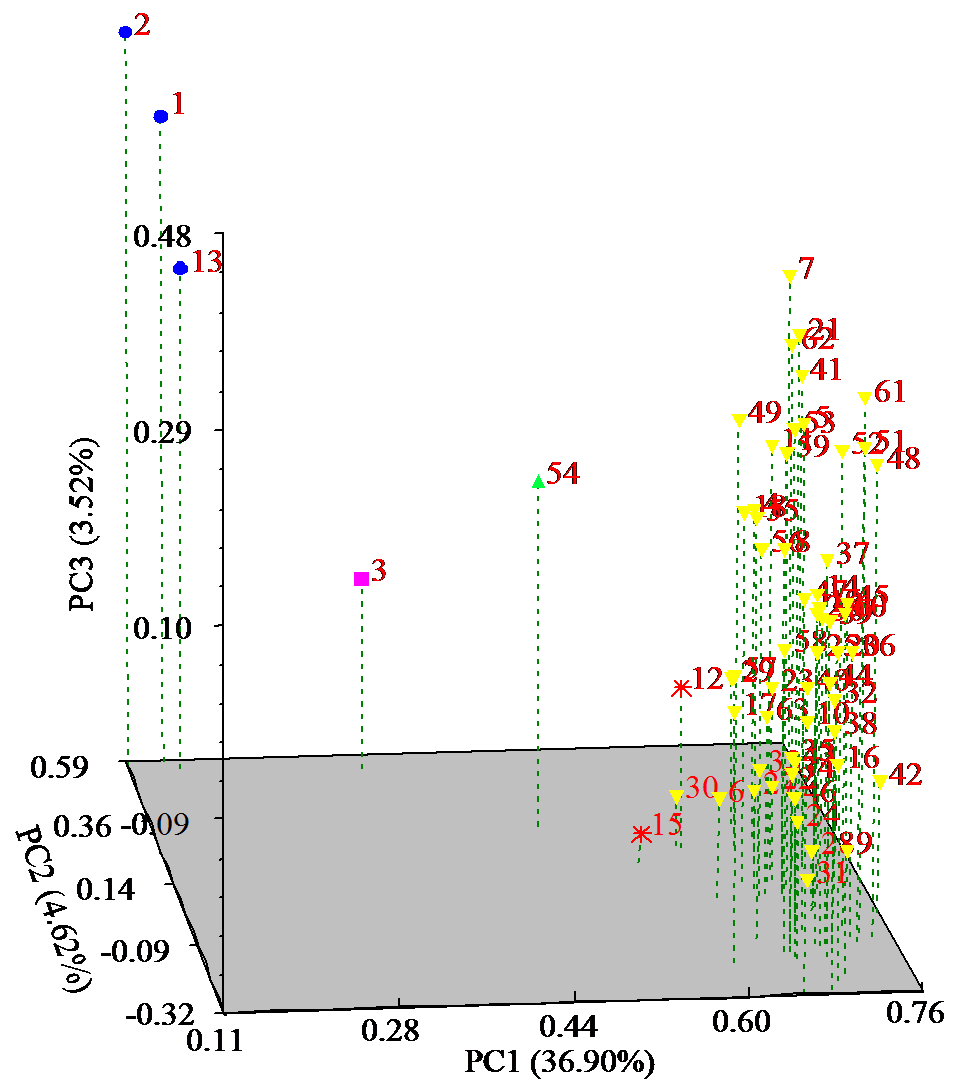
为明确马铃薯种质资源的亲缘关系,利用SSR分子标记对63份马铃薯种质资源的遗传多样性进行分析。结果显示,共扩增到164个多态性条带,其中多态性位点145个,平均条带数为5.13个。有效等位基因数、Nei’s遗传多样性指数和Shannon指数分别为1.7139、0.3927和0.5632,遗传距离范围为0.12~0.71。在遗传距离为0.180处将供试材料分为5个类群,聚类结果与区域高度一致。主成分分析表明,3个主成分的总变异量为45.04%,与聚类结果一致。利用SSR引物STL001、S192和S118构建了63份种质资源的DNA指纹图谱,并建立了二维码分子身份证。本研究为马铃薯资源利用和品种改良奠定基础。
| [1] |
刘思泱, 于卓, 蒙美莲, 等. 6个彩色马铃薯品种的ISSR分析. 华北农学报, 2010, 25(5):117-120.
doi: 10.7668/hbnxb.2010.05.024 |
| [2] | 李文刚, 曹春梅, 刘富强, 等. 国际马铃薯种业现状及发展综述Ⅰ——国际马铃薯种业发展趋势分析. 哈尔滨: 哈尔滨工程大学出版社, 2014. |
| [3] | 杨帅, 闵凡祥, 高云飞. 新世纪中国马铃薯产业发展现状及存在问题. 中国马铃薯, 2014, 28(5):311-316. |
| [4] |
徐建飞, 金黎平. 马铃薯遗传育种研究:现状与展望. 中国农业科学, 2017, 50(6):990-1015.
doi: 10.3864/j.issn.0578-1752.2017.06.003 |
| [5] | 杨学勇, 苏汉东, 张梦卓, 等. 多倍化和驯化研究进展与展望. 中国科学: 生命科学, 2021, 51(10):1457-1466. |
| [6] | 李瑞峰. 甜瓜主栽品种指纹图谱库的构建及遗传多样性分析. 哈尔滨: 东北农业大学, 2014. |
| [7] | 白瑞霞, 彭建营. DNA分子标记在果树遗传育种研究中的应用. 西北植物学报, 2004, 24(8):1547-1554. |
| [8] | 李毳, 李继萍, 柴宝峰. 微卫星标记的发展及植物研究中的应用. 郑州航空工业管理学院学报(社会科学版), 2004, 23(6):199-200. |
| [9] | 李建武, 李灏德, 文国宏, 等. 甘肃省主栽马铃薯品种遗传多样性的AFLP与SSR分子标记分析. 甘肃农业科技, 2016(7):1-6. |
| [10] | 余斌, 杨宏羽, 王丽. 引进马铃薯种质资源在干旱半干旱区的表型性状遗传多样性分析及综合评价. 作物学报, 2018, 44(1):63-74. |
| [11] | 张海雯, 巩檑, 马斯霜. 基于SSR标记的马铃薯种质遗传多样性及群体结构分析. 分子植物育种, 2020, 18(12):4144-4152. |
| [12] | 王鹏, 李芳弟, 郭天顺, 等. 马铃薯种质资源遗传关系分析及指纹图谱构建. 农业生物技术学报, 2020, 28(5):794-810. |
| [13] | 段绍光, 金黎平, 李广存, 等. 马铃薯品种遗传多样性分析. 作物学报, 2017, 43(5):718-729. |
| [14] | Song X Y, Zhang C Z, Li Y, et al. SSR analysis of genetic diversity among 192 diploid potato cultivars. Horticultural Plant Journal, 2016, 2(3):163-171. |
| [15] | Sapinder B, Girijesh P, Rich N, et al. Evaluation of genetic diversity among Russet potato clones and varieties from breeding programs across the United States. PLoS ONE, 2018, 13(8):e0201415. |
| [16] | Karaagac E, Yilma S, Cuesta-Marcos A, et al. Molecular analysis of potatoes from the pacific northwest tri-state variety development program and selection of markers for practical DNA fingerprinting applications. American Journal of Potato Research, 2014, 91(2):195-203. |
| [17] |
Pandey J, Scheuring D C, Koym J W, et al. Genetic diversity and population structure of advanced clones selected over forty years by a potato breeding program in the USA. Scientific Reports, 2021, 11(1):8344.
doi: 10.1038/s41598-021-87284-x pmid: 33863959 |
| [18] | Esnault F, Pelle R, Dantec J P, et al. Development of a potato cultivar core collection, a valuable tool to prospect genetic variation for novel traits. Potato Research, 2016, 59(4):329-343. |
| [19] | Anoumaa M, Yao K N, Kouam B E, et al. Genetic diversity and core collection for potato cultivars from cameroon as revealed by SSR markers. American Journal of Potato Research, 2017, 94(4):449-463. |
| [20] |
McGregor C E, Van T R, Hoekstra R, et al. Analysis of the wild potato germplasm of the series Acaulia with AFLPs: implications for ex situ conservation. Theoretical and Applied Genetics, 2002, 104(1):146-156.
pmid: 12579440 |
| [21] | Moisan-Thiery M, Marhadour S, Kerlan C M, et al. Potato cultivar identification using simple sequence repeats markers (SSR). Potato Research, 2005, 48:191-200. |
| [22] | Galarreta D R I J, Barandalla L, Rios J D, et al. Genetic relationships among local potato cultivars from Spain using SSR markers. Genetic Resources and Crop Evolution, 2011, 58:383-395. |
| [23] | Wang P S, Liu W S, Li Y, et al. Multiplex PCR system optimization with potato SSR markers. Journal of Northeast Agricultural University, 2012, 19(3):20-27. |
| [24] | 贺丹, 唐婉, 刘阳. 尾叶紫薇与紫薇F1代群体主要表型性状与SSR标记的连锁分析. 北京林业大学学报, 2012, 34(6):121-125. |
| [25] | Lee N O, Park Y H. Assessment of genetic diversity incultivated radishes (Raphanus sativus) by agronomic traits and SSR markers. Scientia Horticulturae, 2017, 223:19-30. |
| [26] | Rutuja S P, Krishnan S. Genetic diversity between and within the natural populations of Garcinia indica (Thouars) Choisy: a high value medicinal plant from Northern Western Ghats of India using ISSR markers. Journal of Applied Research on Medicinal and Aromatic Plants, 2019, 15:100219. |
| [27] |
Carneiro L M V, Santini L, Diniz A L, et al. Microsatellite markers: what they mean and why they are so useful. Genetics and Molecular Biology, 2016, 39(3):312-328.
doi: 10.1590/1678-4685-GMB-2016-0027 pmid: 27561112 |
| [28] |
陈小红, 林元香, 王倩, 等. 基于高基元SSR构建黍稷种质资源的分子身份证. 作物学报, 2022, 48(4):908-919.
doi: 10.3724/SP.J.1006.2022.14034 |
| [29] | Singh N, Choudhury R D, Singh K A, et al. Comparison of SSR and SNP markers in estimation of genetic diversity and population structure of Indian rice varieties. PLoS ONE, 2013, 8(12):e84136. |
| [30] | Carvalho M C, Oliveira C M O P, Silva, M S, et al. Genetic diversity and structure of landrace accessions, elite lineages and cultivars of common bean estimated with SSR and SNP markers. Molecular Biology Reports, 2020, 47(9):1-11. |
| [31] | Filippi C V, Aguirre N, Rivas J G, et al. Population structure and genetic diversity characterization of a sunflower association mapping population using SSR and SNP markers. BMC Plant Biology, 2015, 15(1):52. |
| [32] |
Würschum T, Langer S M, Longin C F, et al. Population structure,genetic diversity and linkage disequilibrium in elite winter wheat assessed with SNP and SSR markers. Theoretical and Applied Genetics, 2013, 126(6):1477-1486.
doi: 10.1007/s00122-013-2065-1 pmid: 23429904 |
| [33] | Wu F F, Zhang S X, Gao Q, et al. Genetic diversity and population structure analysis in a large collection of Vicia amoena in China with newly developed SSR markers. BMC Plant Biology, 2021, 21(1):544. |
| [34] | Gil J, Um Y, Kim S, et al. Development of genome-wide SSR markers from Angelica gigas Nakai using next generation sequencing. Genes, 2017, 8(10):238. |
| [35] | Lee H Y, Moon S, Shim D, et al. Development of 44 novel polymorphic SSR markers for determination of shiitake mushroom (Lentinula edodes) cultivars. Genes, 2017, 8(4):109. |
| [36] |
Balbino E, Martins G, Morais S, et al. Genome survey and development of 18 microsatellite markers to assess genetic diversity in Spondias tuberosa Arruda Câmara (Anacardiaceae) and cross-amplification in congeneric species. Molecular Biology Reports, 2019, 46(3):3511-3517.
doi: 10.1007/s11033-019-04768-w pmid: 30915689 |
| [37] | Jayabalan S, Pulipati S, Ramasamy K, et al. Analysis of genetic diversity and population structure using SSR markers and validation of a Cleavage Amplified Polymorphic Sequences (CAPS) marker involving the sodium transporter OsHKT1;5 in saline tolerant rice (Oryza sativa L. ) landraces. Gene, 2019, 713:143976. |
| [38] | Emanuelli F, Lorenzi S, Grzeskowiak L, et al. Genetic diversity and population structure assessed by SSR and SNP markers in a large germplasm collection of grape. BMC Plant Biology, 2013, 13:39. |
| [39] | Chen L, Pan T T, Qian H R, et al. Genetic diversity and population structure revealed by SSR markers on endemic species osmanthusserrulatus rehder from southwestern Sichuan basin, China. Forests, 2022, 12(10):1365. |
| [40] | Sayed M R I, Alshallash K S, Safhi F A, et al. Genetic diversity, analysis of some agro-morphological and quality traits and utilization of plant resources of Alfalfa. Genes, 2022, 13(9):1521. |
| [41] | 潘哲超, 王颖, 徐宁生, 等. 马铃薯重要农艺性状的相关性、主成分与聚类分析. 分子植物育种, 2020, 18(5):1626-1636. |
| [42] | 张招娟, 邓跃, 黄泷健, 等. 福建省马铃薯主栽品种SSR遗传多样性分析. 分子植物育种, 2019, 17(22):7420-7427. |
| [43] | 宋峥, 王崇, 徐颖华, 等. 马铃薯地方种质资源的SSR遗传多样性分析. 分子植物育种, 2020, 20(21):7143-7153. |
| [44] | El-Esawi M A, Germaine K, Bourke P, et al. AFLP analysis of genetic diversity and phylogenetic relationships of Brassica oleracea in Ireland. Comptes Rendus Biologies, 2016, 339(5/6):163-170. |
| [45] | 吴立萍, 吕典秋, 姜丽丽, 等. 俄罗斯马铃薯种质资源遗传多样性的SSR分析. 分子植物育种, 2017, 15(10):4047-4053. |
| [46] | 毛向红, 范向斌, 白小东, 等. 基于SSR分子标记的晋北地区引进马铃薯种质资源遗传多样性分析. 作物杂志, 2024(5):54-59. |
| [47] |
安苗, 王彤彤, 付逸婷, 等. 52个马铃薯遗传多样性分析及SSR分子身份证构建. 生物技术通报, 2023, 39(12):136-147.
doi: 10.13560/j.cnki.biotech.bull.1985.2023-0512 |
| [48] | 张云帅. 24个马铃薯品种的遗传多样性分析及指纹图谱的构建. 张家口: 河北北方学院, 2022. |
| [1] | 高树广, 徐东阳, 胡敏杰, 王瑞霞, 张春花, 徐博涵, 李伟峰, 张留平. 芝麻种质资源品质性状的初步鉴定与评价[J]. 作物杂志, 2025, (4): 58–64 |
| [2] | 孙邦升, 宋继玲, 杨梦平, 邢金月, 胡尊艳, 郝智勇, 李菁华, 刘春生. 高淀粉马铃薯初级核心种质资源的遗传多样性分析[J]. 作物杂志, 2025, (3): 52–60 |
| [3] | 赵雅杰, 温蕊, 贾祎明, 金晓蕾, 张永虎, 张利俊, 张彪, 张慧, 于立霞. 谷子种质资源表型性状遗传多样性分析[J]. 作物杂志, 2025, (3): 61–69 |
| [4] | 常宏兵, 王晨, 何美敬, 曹熙敏, 俞凤芳, 曹晓良, 宋炜, 吕爱枝. 基于SSR标记对69份玉米种质资源的遗传多样性分析[J]. 作物杂志, 2025, (2): 47–53 |
| [5] | 江慧, 钟巧芳, 殷富有, 李金璐, 刘丽, 张云, 王波, 蒋聪, 程在全, 张慧, 肖素勤. 药用野生稻种质资源和多组学研究进展[J]. 作物杂志, 2025, (2): 1–8 |
| [6] | 卢晶, 余波, 江谧, 彭镰心, 任远航, 吴琪. 58份青稞种质资源遗传多样性评价[J]. 作物杂志, 2025, (2): 20–28 |
| [7] | 毛彦芝, 孙贺光, 李庆全, 郭梅, 王文重, 魏琪, 董学志, 闵凡祥, 孙晶, 宋晓宇. 我国马铃薯晚疫病监测预警研究进展[J]. 作物杂志, 2025, (1): 10–14 |
| [8] | 张楷楷, 赵德明, 马菊花, 白鹏军, 马鹏, 陈蕙, 徐文杰, 黄彩霞, 刘众宇. 垄沟秸秆覆盖对旱作马铃薯土壤水热特征及产量的影响[J]. 作物杂志, 2025, (1): 139–146 |
| [9] | 马鹏, 魏熙明, 丁芳菊, 张楷楷, 王巧丽, 邢古月, 常磊, 黄彩霞. 水氮调控对膜下滴灌马铃薯水分动态及产量的影响[J]. 作物杂志, 2025, (1): 170–178 |
| [10] | 颜群翔, 庞玉辉, 洪壮壮, 毕俊鸽, 王春平. 141份国内外小麦种质资源主要性状遗传多样性分析与特异性评价[J]. 作物杂志, 2025, (1): 26–34 |
| [11] | 姚陆铭, 袁娟, 马晓红, 王彪. 基于表型性状及SSR标记的扁豆种质资源遗传多样性分析[J]. 作物杂志, 2025, (1): 35–45 |
| [12] | 张杰, 贾冰, 程瑞宝, 杨薇, 刘影, 张立媛, 温雅辉, 董春浩, 王振普, 琦明玉, 张清艳, 赵敏, 李志光. 东北平原生态区糜子种质资源表型多样性分析[J]. 作物杂志, 2025, (1): 76–82 |
| [13] | 庞敏昡, 王瀚, 李志涛, 史宁帆, 蒲转芳, 张锋, 姚攀锋, 毕真真, 白江平, 孙超. 不同水分处理下施用立收谷对马铃薯品质的影响[J]. 作物杂志, 2024, (6): 132–139 |
| [14] | 阳新月, 向颖, 陈子恒, 林茜, 邓振鹏, 周克友, 李明聪, 王季春. 有机质施用量对马铃薯产量及氮、磷、钾养分吸收利用的影响[J]. 作物杂志, 2024, (6): 153–161 |
| [15] | 金彦君, 祝洪沙, 李成东, 王金禹, 张忠福, 万婷丽, 周爱爱, 撒刚, 程李香, 刘娟, 余斌. 外源激素对雾培马铃薯叶片气孔密度及块茎产量的调控[J]. 作物杂志, 2024, (6): 205–211 |
|
||