Crops ›› 2022, Vol. 38 ›› Issue (4): 14-21.doi: 10.16035/j.issn.1001-7283.2022.04.003
Previous Articles Next Articles
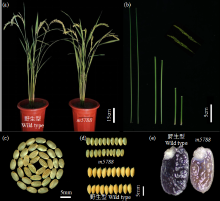
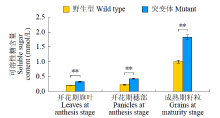
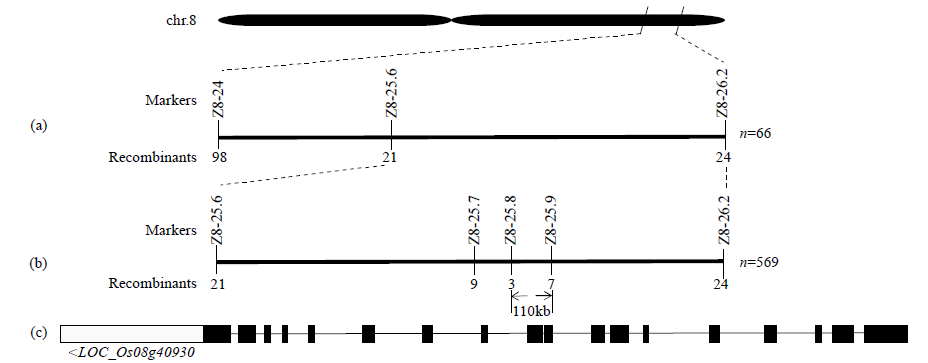
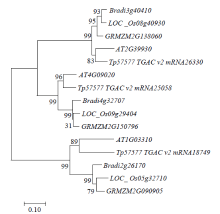
Identification and Fine Mapping of Sugary Endosperm Mutant m5788 in Rice (Oryza sativa L.)
Zheng Siyi1( ), Yang Ye1, Song Yuanhui1, Hua Qin1, Lin Quanxiang1, Zhang Haitao1, Cheng Zhijun2(
), Yang Ye1, Song Yuanhui1, Hua Qin1, Lin Quanxiang1, Zhang Haitao1, Cheng Zhijun2( )
)
- 1College of Agriculture, Anhui Agricultural University, Hefei 230036, Anhui, China
2Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100081, China
| [1] | 金锡铭. 水稻淀粉突变体flo6的表型分析及基因精细定位. 南京:南京农业大学, 2010. |
| [2] |
Manners D J. Recent developments in our understanding of amylopectin structure. Carbohydrate Polymer, 1989, 11(2):87-112.
doi: 10.1016/0144-8617(89)90018-0 |
| [3] |
Jeon J S, Ryoo N, Hahn T R, et al. Starch biosynthesis in cereal endosperm. Plant Physiology Biochemistry, 2010, 48(6):383-392.
doi: 10.1016/j.plaphy.2010.03.006 |
| [4] |
Pfister B, Zeeman S C. Formation of starch in plant cells. Cellular and Molecular Life Sciences, 2016, 73(14):2781-2807.
doi: 10.1007/s00018-016-2250-x |
| [5] |
Dinges J R, Colleoni C, Myers A M, et al. Molecular structure of three mutations at the maize sugary1locus and their allele-specific phenotypic effects. Plant Physiology, 2001, 125:1406-1418.
pmid: 11244120 |
| [6] |
Takahashi S, Kumagai Y, Igarashi H, et al. Biochemical analysis of a new sugary-type rice mutant,Hemi-sugary1,carrying a novel allele of the sugary-1 gene. Planta, 2019, 251(1):1-29.
doi: 10.1007/s00425-019-03297-x |
| [7] |
Sestili F, Sparla F, Botticella E, et al. The down-regulation of the genes encoding Isoamylase 1 alters the starch composition of the durum wheat grain. Plant Science, 2016, 252:230-238.
doi: 10.1016/j.plantsci.2016.08.001 |
| [8] |
Ferreira S J, Senning M, Fischer-Stettler M, et al. Simultaneous silencing of isoamylases ISA1,ISA2and ISA3 by multi-target RNAi in potato tubers leads to decreased starch content and an early sprouting phenotype. PLoS ONE, 2017, 12(7):e0181444.
doi: 10.1371/journal.pone.0181444 |
| [9] |
Peng C, Wang Y H, Liu F, et al. FLOURY ENDOSPERM 6 encodes a CBM 48 domain-containing protein involved in compound granule formation and starch synthesis in rice endosperm. The Plant Journal, 2014, 77(6):917-930.
doi: 10.1111/tpj.12444 |
| [10] | 李家洋, 钱前, 曾大力, 等. 水稻胚乳甜质控制基因SU1及其应用:中国,200510006770.8. 200510006770.8. 2005-02-04. |
| [11] |
赵华, 张其芳, 赵倩, 等. 水稻胚乳糖质突变体Sug-11的淀粉粒结构和粒径分布及相关理化特性. 核农学报, 2015, 29(4):724-733.
doi: 10.11869/j.issn.100-8551.2015.04.0724 |
| [12] |
Du L, Xu F, Fang J, et al. Endosperm sugar accumulation caused by mutation of PHS8/ISA1 leads to pre-harvest sprouting in rice. The Plant Journal, 2018, 95(3):545-556.
doi: 10.1111/tpj.13970 |
| [13] | 张述伟, 宗营杰, 方春燕, 等. 蒽酮比色法快速测定大麦叶片中可溶性糖含量的优化. 食品研究与开发, 2020, 41(7):196-200. |
| [14] |
Kubo A, Colleoni C, Dinges J R, et al. Functions of heteromeric and homomeric isoamylase-type starch-debranching enzymes in developing maize endosperm. Plant Physiology, 2010, 153(3):956-969.
doi: 10.1104/pp.110.155259 |
| [15] |
Streb S, Delatte T, Umhang M, et al. Starch granule biosynthesis in arabidopsis is abolished by removal of all debranching enzymes but restored by the subsequent removal of an endoamylase. Plant Cell, 2008, 20:3448-3466.
doi: 10.1105/tpc.108.063487 |
| [16] |
Dinges J R, Colleoni C, Myers J. Mutational analysis of the pullulanase-type debranching enzyme of maize indicates multiple functions in starch metabolism. Plant Cell, 2003, 15(3):666-680.
pmid: 12615940 |
| [17] |
Li Q F, Zhang G Y, Dong Z W, et al. Characterization of expression of the OsPUL gene encoding a pullulanase-type debranching enzyme during seed development and germination in rice. Plant Physiology Biochemistry, 2009, 47(5):351-358.
doi: 10.1016/j.plaphy.2009.02.001 |
| [18] |
Kawagoe Y, Kubo A, Satoh H, et al. Roles of isoamylase and ADP-glucose pyrophosphorylase in starch granule synthesis in rice endosperm. The Plant Journal, 2005, 42(2):164-174.
doi: 10.1111/j.1365-313X.2005.02367.x |
| [19] |
Kubo A, Fujita N, Harada K, et al. The starch-debranching enzymes isoamylase and pullulanase are both involved in amylopectin biosynthesis in rice endosperm. Plant Physiology, 1999, 121(2):399-409.
pmid: 10517831 |
| [20] |
Fujita N, Toyosawa Y, Utsumi Y, et al. Characterization of pullulanase (PUL)-deficient mutants of rice (Oryza sativa L.) and the function of PUL on starch biosynthesis in the developing rice endosperm. Journal of Experimental Botany, 2009, 60(3):1009-1023.
doi: 10.1093/jxb/ern349 pmid: 19190097 |
| [21] |
赵华, 王俊敏, 张其芳, 等. 水稻糖质胚乳突变体Sug-11籽粒灌浆过程的淀粉合成关键酶活性及其与淀粉理化特性关系. 中国水稻科学, 2015, 29(1):73-81.
doi: 10.3969/j.issn.1001-7216.2015.01.009 |
| [22] | East E M, Hayes H K. Inheritance in maize. Zeitschrift für induktive Abstammungs-und Vererbungslehre, 1911, 6(1):193-196. |
| [23] | 李水琴, 王文瑞, 刘海英, 等. 玉米胚乳遗传基础及相关基因研究. 种子, 2016, 35(6):45-49. |
| [24] |
Nakamura Y, Kubo A, Shimamune T, et al. Correlation between activities of starch debranching enzyme and α-polyglucan structure in endosperms of sugary-1 mutants of rice. The Plant Journal, 1997, 12(1):143-153.
doi: 10.1046/j.1365-313X.1997.12010143.x |
| [25] |
Kubo A, Rahman S, Utsumi Y, et al. Complementation of sugary-1 phenotype in rice endosperm with the wheat isoamylase 1 gene supports a direct role for isoamylase1 in amylopectin biosynthesis. Plant Physiology, 2005, 137(1):43-56.
doi: 10.1104/pp.104.051359 |
|
||