Crops ›› 2022, Vol. 38 ›› Issue (3): 55-62.doi: 10.16035/j.issn.1001-7283.2022.03.008
Previous Articles Next Articles
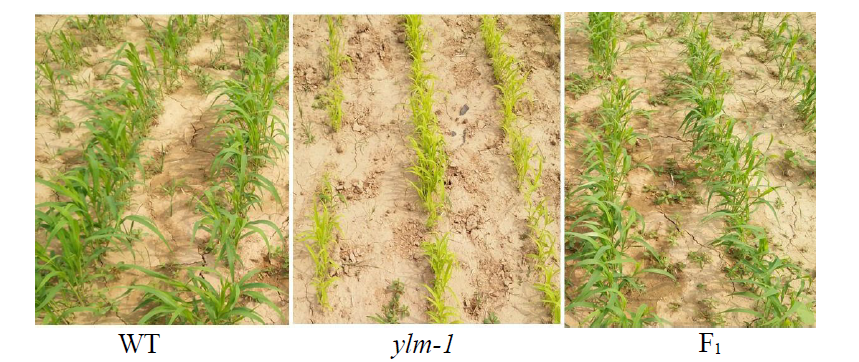
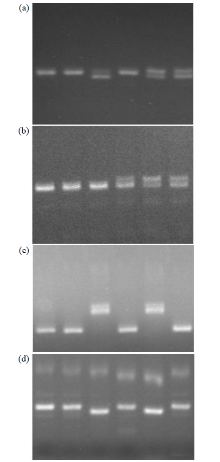
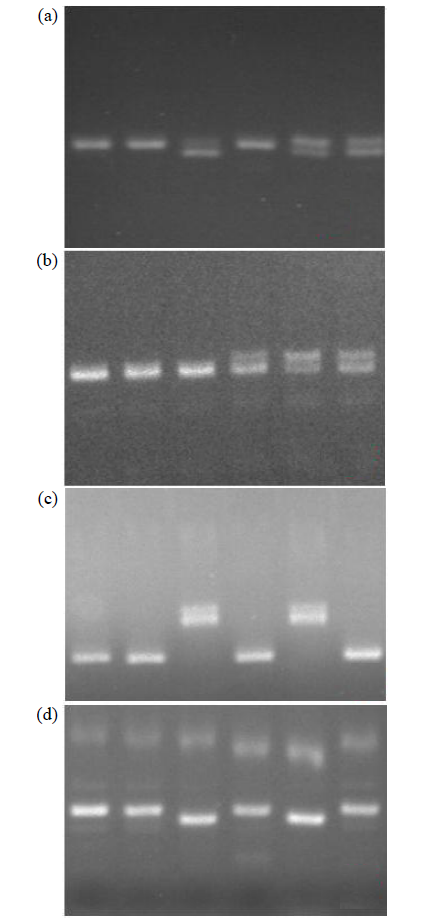
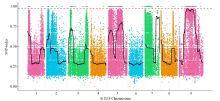
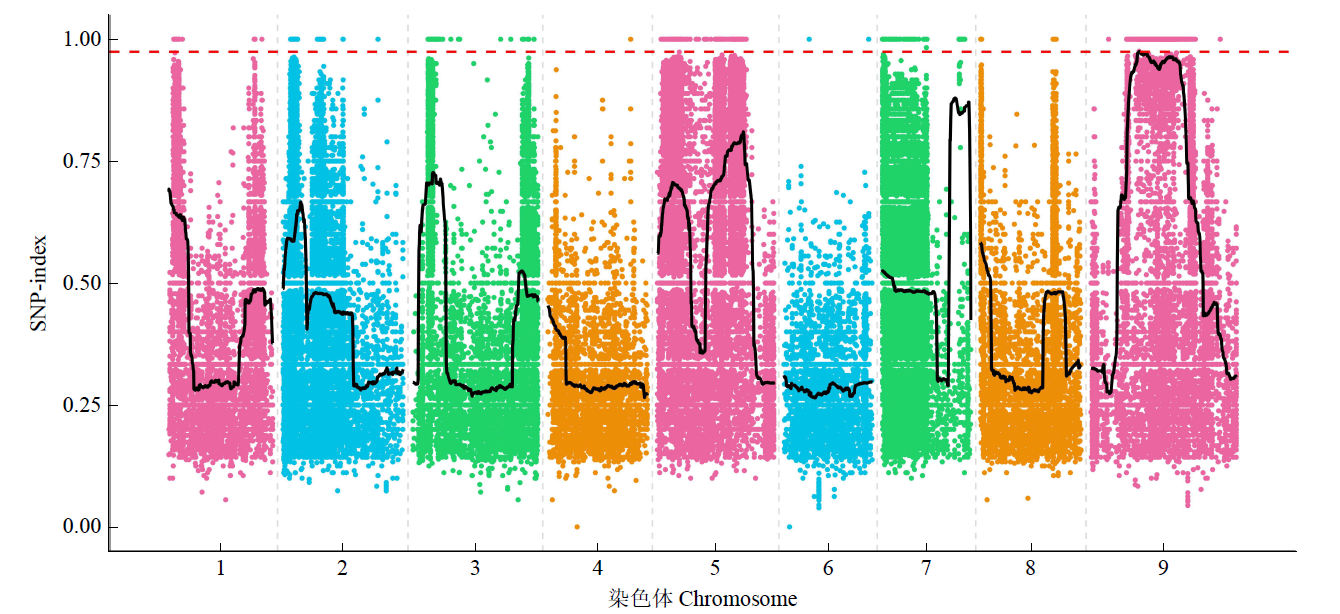
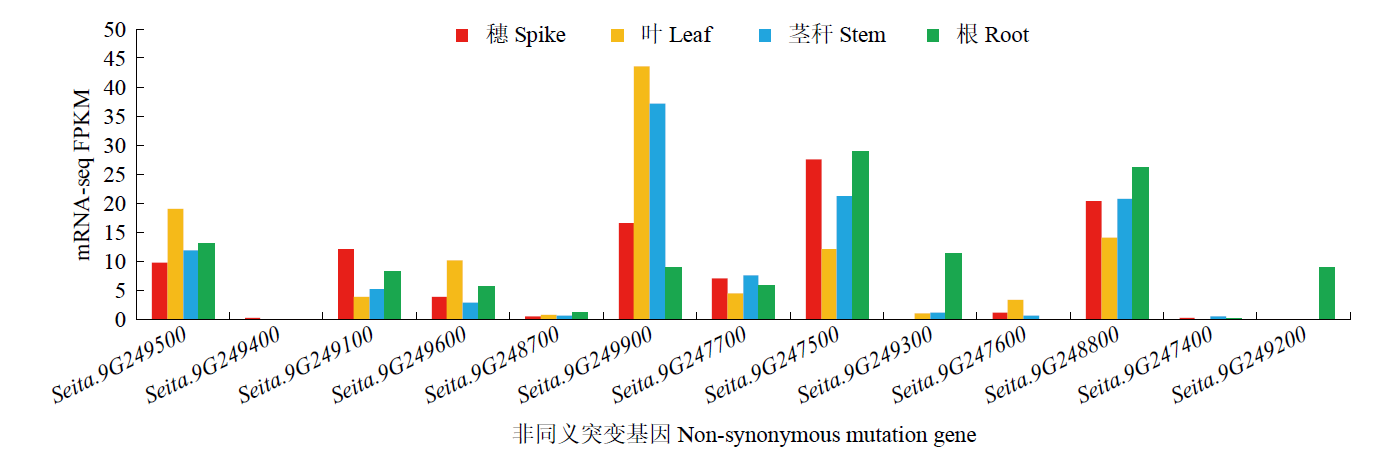
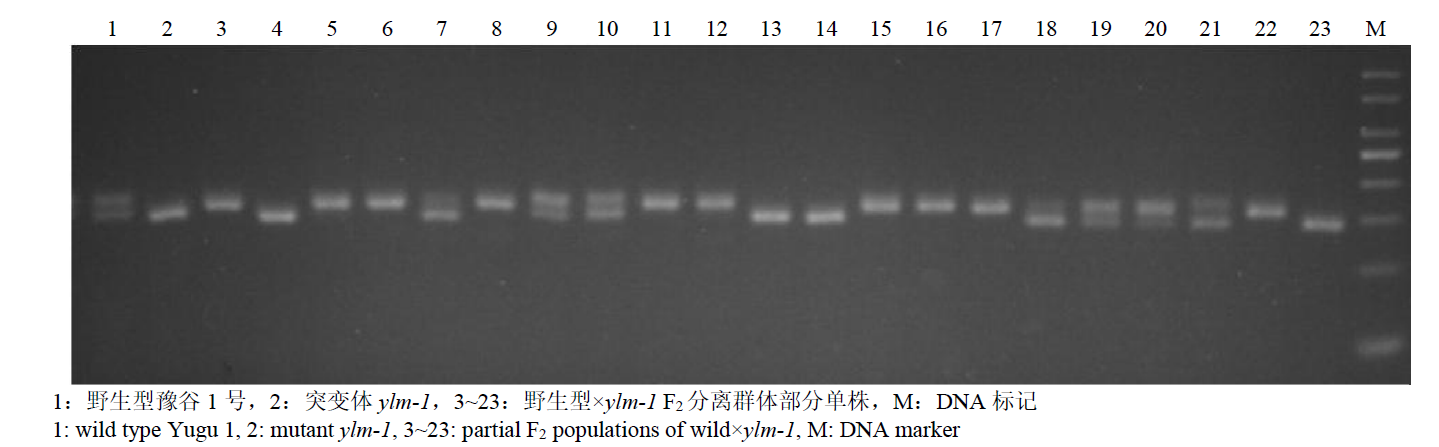
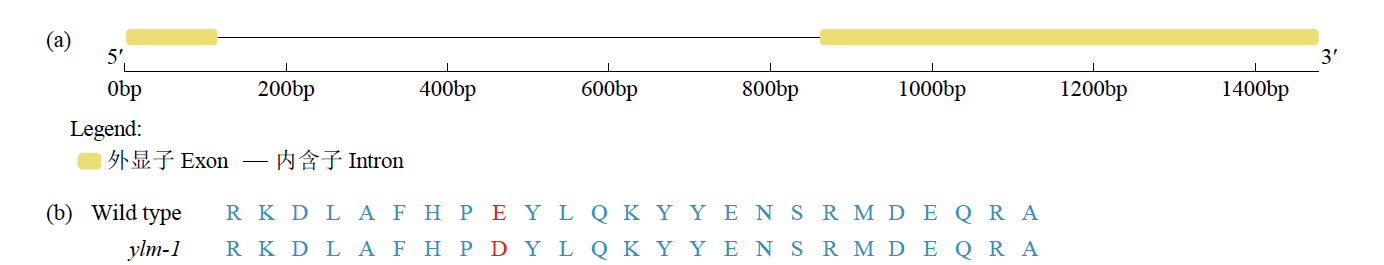
Fine Mapping and Functional Analysis of Yellow Leaf Mutant ylm-1 in Foxtail Millet
Qin Na( ), Zhu Cancan, Dai Shutao, Song Yinghui, Li Junxia(
), Zhu Cancan, Dai Shutao, Song Yinghui, Li Junxia( ), Wang Chunyi
), Wang Chunyi
- Cereal Crops Institute, Henan Academy of Agricultural Sciences, Zhengzhou 450002, Henan, China
| [1] |
Zhao S L, Long W H, Wang Y H, et al. A rice White-stripe leaf (wsl3) mutant lacking an HD domain-containing protein affects chlorophyll biosynthesis and chloroplast development. Journal of Plant Biology, 2016, 59(3):282-292.
doi: 10.1007/s12374-016-0459-8 |
| [2] |
Rooijen R V, Kruijer W, Boesten R, et al. Natural variation of YELLOW SEEDLING 1 affects photosynthetic acclimation of Arabidopsis thaliana. Nature Communications, 2017, 8(1):1424-1432.
doi: 10.1038/s41467-017-01408-4 |
| [3] |
Wang N, Liu Z Y, Zhang Y, et al. Identification and fine mapping of a stay-green gene (Brnye1) in pakchoi (Brassica campestris L. ssp. chinensis). Theoretical and Applied Genetics, 2018, 131(3):673-684.
doi: 10.1007/s00122-017-3028-8 pmid: 29209732 |
| [4] | 邹金财, 张维林, 夏明辉, 等. 水稻阶段性温敏白化转绿突变体stgra254的特征和基因定位. 华北农学报, 2017, 32(3):1-6. |
| [5] |
Junqueira N E G, Ortiz S B, Leal C M V, et al. Anatomy and ultrastructure of embryonic leaves of the C4 species Setaria viridis. Annals of Botany, 2018, 121(6):1163-1172.
doi: 10.1093/aob/mcx217 pmid: 29415162 |
| [6] |
Jia G Q, Huang X H, Zhi H, et al. A haplotype map of genomic variations and genome wide association studies of agronomic traits in foxtail millet (Setaria italica). Nature Genetics, 2013, 45(8):957-961.
doi: 10.1038/ng.2673 |
| [7] | 贾冠清, 刁现民. 谷子(Setaria italica (L. ) P. Beauv.)作为功能基因组研究模式植物的发展现状及趋势. 生命科学, 2017, 29(3):292-301. |
| [8] | 胡彬华, 王平, 杜安平, 等. 水稻淡黄叶突变体pyl3的鉴定和基因定位. 核农学报, 2021, 35(12):2696-2703. |
| [9] | Wang Y, He F, Liu J L, et al. Mapped clone and functional analysis of leaf color gene Ygl7 in a rice hybrid (Oryza sativa L. ssp. indica). PLoS ONE, 2014, 9(6):1-11. |
| [10] | Guan H Y, Xu X B, He C M, et al. Fine mapping and candidate gene analysis of the leaf-color gene ygl-1 in maize. PLoS ONE, 2016, 11(4):1-19. |
| [11] | 李传宗. 谷子苗期黄叶性状的生理基础及候选基因鉴定. 北京: 中国农业科学院, 2020. |
| [12] |
Li W, Tang S, Zhang S, et al. Gene mapping and functional analysis of the novel leaf color gene SiYGL1 in foxtail millet (Setaria italica (L.) P. Beauv). Physiologia Plantarum, 2016, 157(1):24-37.
doi: 10.1111/ppl.12405 |
| [13] |
Tang C J, Tang S, Zhang S, et al. SiSTL1encoding a large subunit of ribonudeotide reductase,is crucial for plant growth,chloroplast biogenesis,and cell cycle progression in Setaria italica. Journal of Experimental Botany, 2019, 70(4):1167-1182.
doi: 10.1093/jxb/ery429 |
| [14] | Zhang S, Tang S, Tang C J, et al. SiSTL2 is required for cell cycle,leaf organ development,chloroplast biogenesis, and has effects on C4 photosynthesis in Setaria italica (L.) P. Beauv. Frontiers in Plant Science, 2018, 9(1):1103-1113. |
| [15] |
Abe A, Kosugi S, Yoshida K, et al. Genome sequencing reveals agronomically important loci in rice using MutMap. Nature Biotechnology, 2012, 30(2):174-178.
doi: 10.1038/nbt.2095 |
| [16] |
Uchida N, Sakamoto T, Kurata T, et al. Identification of EMS induced causal mutations in a non-reference Arabidopsis thaliana accession by whole genome sequencing. Plant and Cell Physiology, 2011, 52(4):716-722.
doi: 10.1093/pcp/pcr029 |
| [17] |
Takagi H, Abe A, Yoshida K, et al. QTL-seq:rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant Journal, 2013, 74(1):174-183.
doi: 10.1111/tpj.12105 |
| [18] |
Tran Q, Bui N, Kappel C, et al. Mapping-by-sequencing via MutMap identifies a mutation in ZmCLE 7 underlying fasciation in a newly developed EMS mutant population in an elite tropical maize inbred. Genes, 2020, 11(3):281.
doi: 10.3390/genes11030281 |
| [19] | 王秋兰, 王智兰, 韩芳, 等. 谷子条纹叶突变体wsl2的鉴定及候选基因分析. 华北农学报, 2020, 35(1):214-221. |
| [20] | 李君霞, 秦娜, 朱灿灿, 等. 谷子黄叶色突变体光合特性研究. 核农学报, 2021, 35(9):1964-1970. |
| [21] | You Q, Zhang L W, Yi X, et al. SIFGD:Setaria italica functional genomics database. Molecular Plant, 2015, 8(6):967-970. |
| [22] |
Liu X P, Yang C, Han F Q, et al. Genetics and fine mapping of a yellow-green leaf gene (ygl-1) in cabbage (Brassica oleracea var. capitata L.). Molecular Breeding, 2016, 36:1-8.
doi: 10.1007/s11032-015-0425-z |
| [23] | James G V, Patel V, Nordström K J, et al. User guide for Mapping by sequencing in Arabidopsis. Genome Biology, 2013, 14(6):61-73. |
| [24] | 李洋洋, 薛冰, 周倩, 等. 黄瓜叶色黄化突变基因yl-2的鉴定. 中国蔬菜, 2020(7):30-37. |
| [25] | 陈竹锋, 严维, 王娜, 等. 利用改进的MutMap方法克隆水稻雄性不育基因. 遗传, 2014, 36(1):85-93. |
| [26] | 袁金红, 李俊华, 袁娇娇, 等. 基于全基因组测序的MutMap方法在正向遗传学研究中的应用. 遗传, 2017, 39(12):1168-1177. |
| [27] |
郭广君, 刁卫平, 刘金兵, 等. 辣椒抗黄瓜花叶病毒病研究进展. 华北农学报, 2014, 29(S1):77-84.
pmid: 19374909 |
|
Sugishima M, Kitamori Y, Noguchi M, et al. Crystal structure of red chlorophyll catabolite reductase:enlargement of the ferredoxin-dependent bilin reductase family. Journal of Molecular Biology, 2009, 389(2):376-387.
doi: 10.1016/j.jmb.2009.04.017 pmid: 19374909 |
|
| [28] |
Pruzinská A, Anders I, Aubry S, et al. In vivo participation of red chlorophyll catabolite reductase in chlorophyll breakdown. Plant Cell, 2007, 19(1):369-387.
pmid: 17237353 |
| [29] |
Xie Z K, Wu S D, Chen J Y, et al. The C‑terminal cysteine-rich motif of NYE1/SGR1 is indispensable for its function in chlorophyll degradation in Arabidopsis. Plant Molecular Biology, 2019, 101(1):257-268.
doi: 10.1007/s11103-019-00902-1 |
| [30] |
Schelbert S, Aubry S, Burla B, et al. Pheophytin pheophorbide hydrolase (pheophytinase) is involved in chlorophyll breakdown during leaf senescence in Arabidopsis. Plant Cell, 2009, 21(3):767-785.
doi: 10.1105/tpc.108.064089 pmid: 19304936 |
| [31] |
Chen Y, Shimoda Y, Yokono M, et al. Mg-dechelatase is involved in the formation of photosystem II but not in chlorophyll degradation in Chlamydomonas reinhardtii. The Plant Journal, 2009, 97(1):1022-1031.
doi: 10.1111/tpj.14174 |
| [32] |
Hauenstein M, Christ B, Das A, et al. A role for TIC 55 as a hydroxylase of phyllobilins,the products of chlorophyll breakdown during plant senescence. Plant Cell, 2016, 28(1):2510-2527.
doi: 10.1105/tpc.16.00630 |
| [33] |
Li Z, Wu S, Chen J, et al. NYEs/SGRs-mediated chlorophyll degradation is critical for detoxification during seed maturation in Arabidopsis. Plant Journal, 2017, 92(1):650-661.
doi: 10.1111/tpj.13710 |
| [34] |
Pruzinská A, Anders I, Aubry S, et al. In vivo participation of red chlorophyll catabolite reductase in chlorophyll breakdown. Plant Cell, 2007, 19(1):369-387.
pmid: 17237353 |
| [35] |
Fukasawa A, Suzuki Y, Terai H, et al. Effects of postharvest ethanol vapor treatment on activities and gene expression of chlorophyll catabolic enzymes in broccoli florets. Postharvest Biology and Technology, 2010, 55(2):97-102.
doi: 10.1016/j.postharvbio.2009.08.010 |
| [36] |
Gomez-Lobato M E, Civello P M, Martinez G A. Effects of ethylene,cytokinin and physical treatments on BoPaO gene expression of harvested broccoli. Journal of the Science of Food and Agriculture, 2012, 92(1):151-158.
doi: 10.1002/jsfa.4555 pmid: 21732385 |
| [1] | Zheng Siyi, Yang Ye, Song Yuanhui, Hua Qin, Lin Quanxiang, Zhang Haitao, Cheng Zhijun. Identification and Fine Mapping of Sugary Endosperm Mutant m5788 in Rice (Oryza sativa L.) [J]. Crops, 2022, 38(4): 14-21. |
| [2] | Ma Ke, Feng Lei, Zhao Xiatong, Zhang Liguang, Yuan Xiangyang, Dong Shuqi, Guo Pingyi, Song Xi’e. Effects of Sowing Distance and Sowing Amount on the Growth Characteristics and Yield of Zhangzagu 10 [J]. Crops, 2022, 38(4): 172-178. |
| [3] | Lü Jianzhen, Ren Ying, Wang Hongyong, Zhang Tingjun, Ma Jianping, Zhao Kai. Comprehensive Phenotype Evaluation of 264 Major Foxtail Millet Bred Varieties (Lines) [J]. Crops, 2022, 38(4): 22-31. |
| [4] | Li Binghua, Wang Guiqi, Shi Zhigang, Liu Xiaomin, Xu Xian, Zhao Bochui, Cheng Ruhong. Sensitivity of Foxtail Millets (Setaria italica L.) and Weeds to Cyhalofop-Butyl [J]. Crops, 2022, 38(4): 262-266. |
| [5] | Guo Yongxin, Zhou Hao, Sun Peng, Wang Yaqing, Ma Ke, Li Xiaorui, Dong Shuqi, Guo Pingyi, Yuan Xiangyang. Effects of Planting Patterns on Lodging Resistance and Yield of Zhangza 10 in Different Ecological Areas [J]. Crops, 2022, 38(2): 195-202. |
| [6] | Zhao Lirong, Ma Ke, Zhang Liguang, Tang Sha, Yuan Xiangyang, Diao Xianmin. Analysis of Agronomic Traits and Quality of Foxtail Millet Varieties in Different Ecological Regions [J]. Crops, 2022, 38(2): 44-53. |
| [7] | Li Huixia, Liu Hong, Wang Yuwen, Tian Gang, Liu Xin, Zheng Zhiyin. Study on the Technique of Removing False Hybrids from Foxtail Millet Herbicide-Resistant Hybrids [J]. Crops, 2021, 37(6): 72-77. |
| [8] | Wang Yuting, Miao Xingfen, Wang Di. Screening and Evaluation of Atrazine-Resistant Germplasm Resources of Foxtail Millet at Germination Stage [J]. Crops, 2021, 37(5): 194-204. |
| [9] | Gao Peng, Guo Meijun, Yang Xuefang, Dong Shuqi, Wen Yinyuan, Guo Pingyi, Yuan Xiangyang. Responses of Photosynthetic Fluorescence Parameters in Foxtail Millet and Maize Leaves under Nicosulfuron Stress [J]. Crops, 2021, 37(3): 70-77. |
| [10] | Shen Jie, Wang Yuguo, Guo Pingyi, Yuan Xiangyang. Effects of Humic Acid on Ascorbate-Glutathione Cycle in the Leaves of Foxtail Millet Seedlings under Drought Stress [J]. Crops, 2021, 37(2): 173-177. |
| [11] | Jia Suqing, He Lu, Du Yanwei. Effects of Different Tillage Methods on Root Development,Yield and Water Use Efficiency of Spring Millet in Arid Area [J]. Crops, 2020, 36(5): 194-198. |
| [12] | Yang Yongqing, Gao Fangfang, Ma Yajun, Chen Xin, Zhang Jie. Effects of Different Fertilizer Treatments on Yield, Quality and Economic Benefit of Foxtail Millet in Dry Farming Area of Shanxi Province [J]. Crops, 2020, 36(4): 195-201. |
| [13] | Zhao Lijuan,Yuan Hongmei,Zhao Liwei,Guo Wendong,Li Zhijiang,Li Xiangyu,Ma Jinfeng,Li Yandong,Song Weifu,Yang Xuefeng,Liu Dongjun. The Phenotypic Variations and GA Sensitivity of a Dwarf Mutant d93090 in Foxtail Millet [J]. Crops, 2019, 35(6): 27-32. |
| [14] | Zhang Di,Miao Xingfen,Wang Yuting. Evaluation and Screening of Salt Tolerance in 100 Foxtail Millet at Germination Stage [J]. Crops, 2019, 35(6): 43-49. |
| [15] | Yue Linqi,Shi Weiping,Guo Jiahui,Guo Pingyi,Guo Jie. Response of Cutin Synthetic Genes of Foxtail Millet to Drought Stress [J]. Crops, 2019, 35(4): 183-190. |
|
||