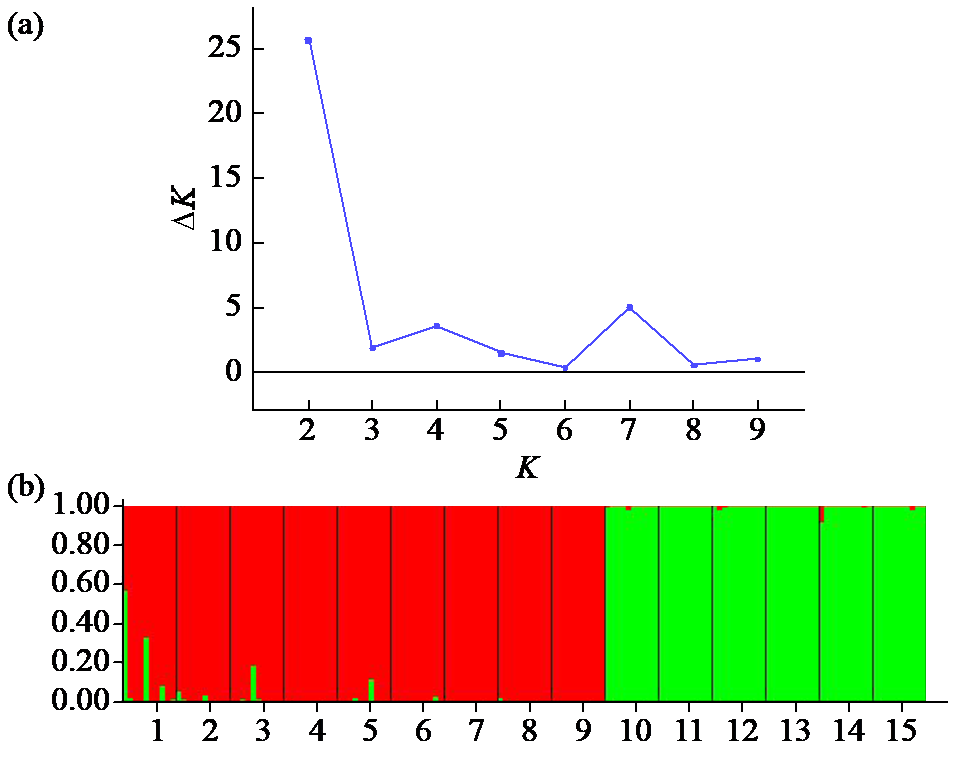
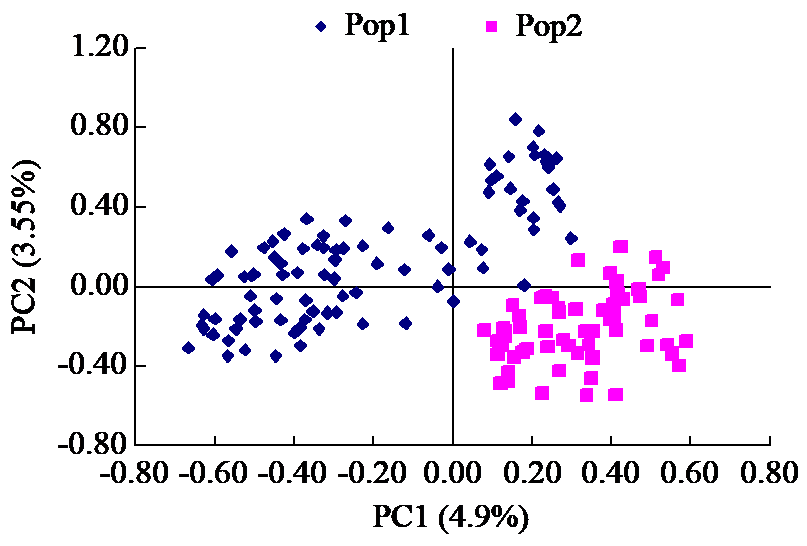
Crops ›› 2023, Vol. 39 ›› Issue (1): 30-37.doi: 10.16035/j.issn.1001-7283.2023.01.005
Previous Articles Next Articles
Genetic Diversity Analysis of Sophora flavescens Ait. Germplasm Resources Based on cpSSR Markers
Song Yun1,2( ), Zhang Xinrui1, He Jiaxin1, Li Zheng1, Sun Zhe1, Li Aoxuan1, Qiao Yonggang1,2(
), Zhang Xinrui1, He Jiaxin1, Li Zheng1, Sun Zhe1, Li Aoxuan1, Qiao Yonggang1,2( )
)
- 1College of Life Sciences, Shanxi Agricultural University, Jinzhong 030801, Shanxi, China
2Shanxi Key Laboratory for Modernization of TCVM, Jinzhong 030801, Shanxi, China
| [1] | 中国科学院《中国植物志》编辑委员会. 中国植物志. 北京: 科学出版社, 1994. |
| [2] | 国家药典委员会. 中国人民共和国药典(一部). 北京: 中国医药科技出版社, 2020:211. |
| [3] | 张晓雯, 李凌宇, 尚海, 等. 苦参碱及其类似物的结构修饰研究进展. 中草药, 2019, 50(23):5892-5900. |
| [4] | 葛淑俊, 孟义江, 李广敏, 等. 我国药用植物遗传多样性研究进展. 中草药, 2006(10):1584-1589. |
| [5] |
Li Q, Su X, Ma H, et al. Development of genic SSR marker resources from RNA-seq data in Camellia japonica and their application in the genus Camellia. Scientific Reports, 2021, 11(1):9919.
doi: 10.1038/s41598-021-89350-w |
| [6] |
Grover A, Sharma P C. Development and use of molecular markers:past and present. Critical Reviews in Biotechnology, 2016, 36(2):290-302.
doi: 10.3109/07388551.2014.959891 pmid: 25430893 |
| [7] | 张征锋, 肖本泽. 基于生物信息学与生物技术开发植物分子标记的研究进展. 分子植物育种, 2009, 7(1):130-136. |
| [8] | 张全芳, 姜明松, 陈峰, 等. 山东省水稻品种(系)的遗传多样性分析. 作物杂志, 2021(4):26-31. |
| [9] | 李琼, 常世豪, 武婷婷, 等. 120份大豆种质资源遗传多样性和亲缘关系分析. 作物杂志, 2021(4):51-58. |
| [10] |
Tonti-Filippini J, Nevill P G, Dixon K, et al. What can we do with 1000 plastid genomes?. The Plant Journal, 2017, 90(4):808-818.
doi: 10.1111/tpj.13491 pmid: 28112435 |
| [11] |
Zhou J, Chen X, Cui Y, et al. Molecular structure and phylogenetic analyses of complete chloroplast genomes of two aristolochia medicinal species. International Journal of Molecular Sciences, 2017, 18(9):1839.
doi: 10.3390/ijms18091839 |
| [12] |
Yu X Q, Drew B T, Yang J B, et al. Comparative chloroplast genomes of eleven Schima (Theaceae) species:insights into DNA barcoding and phylogeny. PLoS ONE, 2017, 12:e0178026.
doi: 10.1371/journal.pone.0178026 |
| [13] | Xu C, Dong W P, Li W Q, et al. Comparative analysis of six Lagerstroemia complete chloroplast genomes. Frontiers in Plant Science, 2017, 8:15. |
| [14] |
Gu C H, Tembrock L R, Zheng S Y, et al. The complete chloroplast genome of Catha edulis:a comparative analysis of genome features with related species. International Journal of Molecular Sciences, 2018, 19(2):525.
doi: 10.3390/ijms19020525 |
| [15] | 杜久军, 左力辉, 刘易超, 等. 裂叶榆叶绿体基因组及CP-SSR位点分析. 植物遗传资源学报, 2018, 19(6):1187-1196. |
| [16] | 薛宏, 易自力, 肖亮, 等. 芒属植物叶绿体InDel标记的开发与应用. 现代农业科技, 2015(7):153-155,159. |
| [17] | Zhang S A, Gao M Q, Zaitlin D. Molecular linkage mapping and marker-trait associations with NIRPT,a downy mildew resistance gene in Nicotiana langsdorffii. Frontiers in Plant Science, 2012, 3:185. |
| [18] |
Breidenbach N, Gailing O, Krutovsky K V. Genetic structure of coast redwood (Sequoia sempervirens [D. Don] Endl.) populations in and outside of the natural distribution range based on nuclear and chloroplast microsatellite markers. PLoS ONE, 2020, 15 (12):e0243556.
doi: 10.1371/journal.pone.0243556 |
| [19] |
Gichira A W, Avoga S, Li Z Z, et al. Comparative genomics of 11 complete chloroplast genomes of Senecioneae (Asteraceae) species:DNA barcodes and phylogenetics. Botanical Studies, 2019, 60(1):17.
doi: 10.1186/s40529-019-0265-y |
| [20] | 高源, 王大江, 王昆, 等. 新疆野苹果叶绿体DNA变异与遗传进化分析. 植物遗传资源学报, 2020, 21(3):579-587. |
| [21] |
Aecyo P, Marques A, Huettel B, et al. Plastome evolution in the Caesalpinia group (Leguminosae) and its application in phylogenomics and populations genetics. Planta, 2021, 254(2):27.
doi: 10.1007/s00425-021-03655-8 pmid: 34236509 |
| [22] | 张靖国, 曹玉芬, 陈启亮, 等. 基于叶绿体DNA变异的湖北梨属种质系统进化及遗传多样性分析. 植物遗传资源学报, 2016, 17(4):766-772. |
| [23] | 王荣升, 杨庆文. 基于叶绿体基因多样性的中国水稻起源进化研究. 植物遗传资源学报, 2011, 12(5):686-693. |
| [24] |
Shahzadi I, Ahmed R, Hassan A, et al. Optimization of DNA extraction from seeds and fresh leaf tissues of wild marigold (Tagetes minuta) for polymerase chain reaction analysis. Genetics and Molecular Research, 2010, 9(1):386-393.
doi: 10.4238/vol9-1gmr747 pmid: 20309824 |
| [25] | 乔永刚, 贺嘉欣, 王勇飞, 等. 药用植物苦参的叶绿体基因组及其特征分析. 药学学报, 2019, 54(11):2106-2112. |
| [26] |
Beier S, Thiel T, Münch T, et al. MISA-web:a web server for microsatellite prediction. Bioinformatics, 2017, 33(16):2583-2585.
doi: 10.1093/bioinformatics/btx198 |
| [27] |
Peakall R, Smouse P E. GenAlEx 6.5:genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics, 2012, 28(19):2537-2539.
pmid: 22820204 |
| [28] |
Liu K J, Muse S V. Power Marker:an integrated analysis environment for genetic marker analysis. Bioinformatics, 2005, 21(9):2128-2129.
doi: 10.1093/bioinformatics/bti282 |
| [29] |
Kumar S, Stecher G, Li M, et al. MEGA X:molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 2018, 35(6):1547-1549.
doi: 10.1093/molbev/msy096 |
| [30] |
Saitou N, Nei M. The neighbor-joining method:a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution, 1987, 4(4):406-425.
doi: 10.1093/oxfordjournals.molbev.a040454 pmid: 3447015 |
| [31] |
Evanno G, Regnaut S, Goudet J. Detexting the number of clusters of individuals using the software STRUCTURE:a simulation study. Molecular Ecology, 2005, 14(8):2611-2620.
doi: 10.1111/j.1365-294X.2005.02553.x pmid: 15969739 |
| [32] |
Earl D A, Vonholdt B M. STRUCTURE HARVESTER:a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources, 2012, 4(2):359-361.
doi: 10.1007/s12686-011-9548-7 |
| [33] | 王崇, 王连军, 杨新笋, 等. 104个甘薯品种的cpSSR指纹图谱构建及遗传多样性分析. 热带作物学报, 2021, 42(6):1549-1556. |
| [34] | 魏潇, 章秋平, 刘威生, 等. 基于叶绿体SSR单倍型的普通杏演化关系. 植物遗传资源学报, 2018, 19(4):705-712. |
| [35] | Yan Y D, Li X Y, Worth J R P, et al. Development of chloroplast microsatellite markers for Glyptostrobus pensilis (Cupressaceae). Applications in Plant Sciences, 2019, 7(7):e11277. |
| [36] | 李祥栋, 石明, 陆秀娟, 等. 利用叶绿体基因组SSR标记揭示薏苡属种质资源的遗传多样性. 华北农学报, 2019, 34(增1):6-14. |
| [37] | 段永红, 渠云芳, 王长彪, 等. 药用植物苦参SSR-PCR体系的优化与验证. 中国农业大学学报, 2014, 19(5):95-100. |
| [38] |
刘亚令, 耿雅萍, 解潇冬, 等. 基于SSR分子标记的药用黄芪遗传多样性与遗传结构分析. 草地学报, 2019, 27(5):1154-1162.
doi: 10.11733/j.issn.1007-0435.2019.05.006 |
| [39] |
程丽莉, 胡广隆, 苏淑钗, 等. 板栗及其近缘种叶绿体SSR遗传多样性分析. 华北农学报, 2015, 30(2):145-149.
doi: 10.7668/hbnxb.2015.02.026 |
| [1] | Huang Guibin, Guan Yaobing, Niu Yongqi, Zhou Lilei, Zhao Yongfeng. Comprehensive Evaluation of 12 Major Agronomic Traits of 103 Chickpea Germplasm Resources [J]. Crops, 2023, 39(1): 6-13. |
| [2] | Guo Huanle, Tang Bin, Li Han, Cao Zhongyang, Zeng Qiang, Liu Liangwu, Chen Zhihui. Comprehensive Evaluation of Phenotypic Traits and Classification of Maize Landraces in Hunan Province [J]. Crops, 2022, 38(6): 33-41. |
| [3] | Li Wangsheng, Wang Xueqian, Yin Xilong, Shi Yang, Liu Dali, Tan Wenbo, Xing Wang. Comprehensive Evaluation of Drought Tolerance of Sugar Beet Germplasms at Seedling Stage [J]. Crops, 2022, 38(6): 54-60. |
| [4] | Zhao Xiaoqin, Jia Ruiling, Liu Junxiu, Liu Yanming, Wen Yinhua, Shi Lili, Zhang Juanning, Ma Ning. Agronomic Traits and Genetic Diversity Analysis of 120 Foxtail Millet Germplasms [J]. Crops, 2022, 38(6): 61-69. |
| [5] | Qi Guangxun, Dong Lingchao, Zhang Wei, Yuan Cuiping, Liu Xiaodong, Wang Yingnan, Dong Yingshan, Wang Yumin, Zhao Hongkun. Evaluation of Resistance to Soybean Mosaic Virus Strain 3 (SMV3) in Foreign Soybean Germplasm Resources [J]. Crops, 2022, 38(6): 70-74. |
| [6] | Lei Lei, Guan Zheyun, Cao Shiliang, Wang Yumin, Lin Chunjing, Peng Bao, Liu Peng, Zhao Limei, Li Zhigang, Zhang Chunbao. Classification of Soybean Heterotic Groups Based on SSR Molecular Markers for Yield-Related Traits [J]. Crops, 2022, 38(4): 54-61. |
| [7] | Zhao Yanfei, Wang Jiyong. Germplasm Resource Protection and Breeding Innovation Status and Development Countermeasures——Taking Hunan and Hainan Provinces as Examples [J]. Crops, 2022, 38(2): 1-5. |
| [8] | Li Wenlue, Chen Changli, Luo Xiahong, Liu Tingting, An Xia, Jin Guanrong, Zhu Guanlin. Genetic Diversity Analysis of Phenotypic Characteristics of Kenaf Resources in Zhejiang Province [J]. Crops, 2022, 38(1): 50-55. |
| [9] | Gao Zhanning, Wang Shujie, Feng Hui, Xue Zhenggang, Yang Yongqian, Song Xiaopeng, Jie Yuanfen. Comprehensive Evaluation of Two-Rowed Barley Varieties (Lines) [J]. Crops, 2022, 38(1): 70-76. |
| [10] | Zhang Quanfang, Jiang Mingsong, Chen Feng, Zhu Wenyin, Zhou Xuebiao, Yang Lianqun, Xu Jiandi. Analysis of Genetic Diversity of Rice Varieties (Lines) in Shandong Province [J]. Crops, 2021, 37(4): 26-31. |
| [11] | Li Qiong, Chang Shihao, Wu Tingting, Geng Zhen, Yang Qingchun, Shu Wentao, Li Jinhua, Zhang Donghui, Zhang Baoliang. Analysis of Genetic Diversity and Genetic Relationship for 120 Soybean Germplasms [J]. Crops, 2021, 37(4): 51-58. |
| [12] | Jia Ruiling, Zhao Xiaoqin, Nan Ming, Chen Fu, Liu Yanming, Wei Liping, Liu Junxiu, Ma Ning. Genetic Diversity Analysis and Comprehensive Assessment of Agronomic Traits of 64 Tartary Buckwheat Germplasms [J]. Crops, 2021, 37(3): 19-27. |
| [13] | Xiang Chao, Sun Suli, Zhu Zhendong, Zong Xuxiao, Yang Tao, Liu Rong, Yang Mei, Xian Dongfeng, Yang Xiuyan. Resistance and Molecular Identification to Powdery Mildew of Pea Germplasms in Sichuan [J]. Crops, 2021, 37(3): 51-56. |
| [14] | Pan Xiaoxue, Hu Mingyu, Wang Zhongwei, Wu Hong, Lei Kairong. Evaluation of Agronomic Traits and Cold Tolerance at Germination Stage in Rice (Oryza sativa L.) Germplasms [J]. Crops, 2021, 37(1): 47-53. |
| [15] | Yang Wanjun, Pan Xiangyu, Wang Xiuhua, Wang Lu, Zhao Yan. Genetic Diversity Analysis of Yield and Agronomic Traits of 119 Alfalfa Varieties (Lines) [J]. Crops, 2020, 36(6): 17-22. |
|
||