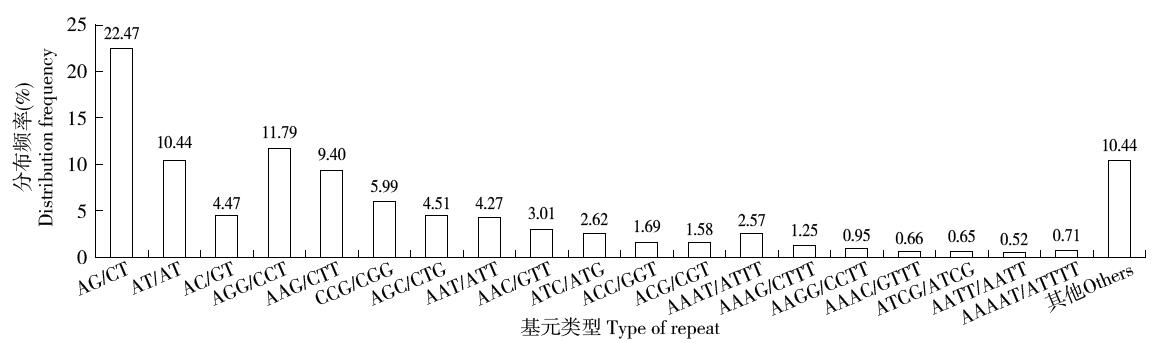
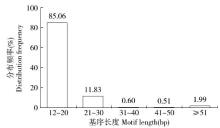
Crops ›› 2016, Vol. 32 ›› Issue (3): 171-174.doi: 10.16035/j.issn.1001-7283.2016.03.032
SSR Information in Transcriptome of White Ginger (Zingiber officinale Roscoe)
Zou Yong1,2,Huang Ke1,2,Jiang Yusong1,2,Liu Yiqing1,2
- 1Forestry and Life Sciences,Chongqing University of Sciences and Arts,Chongqing 402160,China
2Engineering Research Center for Special Plant Seedlings of Chongqing,Chongqing 402160,China
| [1] |
Zhang X H, Liu T J, Li X X , et al. Interspecific hybridization,polyploidization,and backcross of Brassica oleracea var.alboglabra with B.rapa var.purpurea morphologically recapitulate the evolution of Brassica vegetables. Scientific Reports, 2016,6:18618.
doi: 10.1038/srep18618 |
| [2] | 胡建斌, 刘颖, 王兰菊 , 等. 甜瓜EST序列中微卫星的分布特征. 植物生理学通讯, 2009,45(3):258-262. |
| [3] | Wang J Y, Liang Y L, Hai M R , et al. Genome-Wide transcriptional excavation of Dipsacus asperoides unmasked both cryptic asperosaponin biosynthetic genes and SSR markers. Frontiers in Plant Science, 2016,7:339. |
| [4] |
张庆田, 李晓艳, 杨义明 , 等. 蓝靛果忍冬转录组SSR信息分析及其分子标记开发. 园艺学报, 2016,43(3):557-563.
doi: 10.16420/j.issn.0513-353x.2015-0659 |
| [5] |
段紫英, 胡丽芳, 黄长干 , 等. 黄瓜EST中SSR的分布特征分析. 广东农业科学, 2012,39(22):160-162.
doi: 10.3969/j.issn.1004-874X.2012.22.052 |
| [6] |
李满堂, 张仕林, 邓鹏 , 等. 洋葱转录组SSR信息分析及其多态性研究. 园艺学报, 2015,42(6):1103-1111.
doi: 10.16420/j.issn.0513-353x.2015-0046 |
| [7] | 李炎林, 杨星星, 张家银 , 等. 南方红豆杉转录组SSR挖掘及分子标记的研究. 园艺学报, 2014,41(4):735-745. |
| [8] | 张得芳, 李淑娴, 夏涛 .蔷薇科6个属植物EST-SSR特征分析.植物研究,2014(6):810-815. |
| [9] |
王森, 张震, 姜倪皓 , 等.半夏转录组中的SSR位点信息分析.中药材,2014(9):1566-1569.
doi: 10.13863/j.issn1001-4454.2014.09.015 |
| [10] |
鄢秀芹, 鲁敏, 安华明 . 刺梨转录组SSR信息分析及其分子标记开发. 园艺学报, 2015,42(2):341-349.
doi: 10.16420/j.issn.0513-353x.2014-0882 |
| [11] | 程小毛, 宗媛媛, 黄晓霞 .挪威云杉EST资源的SSR信息分析.现代农业科技,2011(8):185-186. |
| [12] |
伍宝朵, 郝朝运, 范睿 , 等. 胡椒EST-SSR标记的开发和应用. 热带作物学报, 2015,36(2):237-243.
doi: 10.3969/j.issn.1000-2561.2015.02.004 |
| [13] | 黄益洪, 张强, 张大勇 , 等. 海蓬子属植物EST序列的SSR信息分析与标记开发. 江苏农业学报, 2013,29(6):1271-1277. |
| [1] | Tang Liyuan, Li Xinghe, Zhang Sujun, Wang Haitao, . QTL Mapping for Photosynthesis#br# Related Traits in Upland Cotton [J]. Crops, 2018, 34(5): 85-90. |
| [2] | Huiyao Tian,Jizhi Jiang,Chengbin Li,Fen Shen,Ning Hou. Genetic Diversity of Phytophthora infestans in Northeast China [J]. Crops, 2018, 34(3): 168-173. |
| [3] | Rui Li,Jianrong Bai,Xiuhong Wang,Congzhuo Zhang,Xiaomei Zhang,Lei Yan,Ruijuan Yang. Population Genetic Diversity of 144 Sweet Maizes [J]. Crops, 2018, 34(2): 17-24. |
| [4] | Shuai Zhang,Yuhui Pang,Zhenghong Wang,Liming Wang,Chunyan Chen,Zhankui Zeng,Chunping Wang. Variation of Agronomic Traits and Genetic Diversity in Wheat Germplasms [J]. Crops, 2018, 34(2): 44-51. |
| [5] | Shanshan Lu,Chenglai Wu,Yan Li,Chunqing Zhang. The Molecular Basis of Holding the Feature and Genetic Purity for Maize Inbred Lines [J]. Crops, 2018, 34(1): 41-48. |
| [6] | Lihua Liu,Shaohua Yuan,Shuying Feng,Binshuang Pang,Hongbo Li,Yangna Liu,Liping Zhang,Changping Zhao. Genetic Difference Analysis and Construction of SSR Fingerprinting Database for F-Type Wheat Male Sterile Line and Restorer Lines [J]. Crops, 2017, 33(6): 30-36. |
| [7] | Lianmei Tai,Yuhu Zuo,Yaling Zhang,Yongling Jin,Yongxia Guo,Xuehui Jin,Guanghui Jin. Analysis of Genetic Diversity of Alternaria solani Isolated from Potato in Heilongjiang Province [J]. Crops, 2017, 33(3): 151-156. |
| [8] | Lihua Liu,Binshuang Pang,Hongbo Li,Yangna Liu,Deping Fu,Na Wang,Changping Zhao. SSR Analysis of Winter Wheat Lines of Beijing Regional Trials from 2009 to 2015 [J]. Crops, 2016, 32(5): 13-18. |
| [9] | Sujun Zhang,Liyuan Tang,Cunjing Liu,Zhenxing Jiang,Jina Chi,Haiyan Tian,Xinghe Li,Jianhong Zhang,Xiangyun Zhang. Association Analysis of Fiber Quality with SSR Markers in Gossypium barbadense L. [J]. Crops, 2016, 32(4): 93-100. |
|