作物杂志,2021, 第2期: 35–44 doi: 10.16035/j.issn.1001-7283.2021.02.005
所属专题: 油料作物
花生TCP转录因子的全基因组鉴定及组织表达特性分析
王通1( ), 赵孝东2, 甄萍萍3, 陈静1, 陈明娜1, 陈娜1, 潘丽娟1, 王冕1, 许静1, 禹山林1, 迟晓元1(
), 赵孝东2, 甄萍萍3, 陈静1, 陈明娜1, 陈娜1, 潘丽娟1, 王冕1, 许静1, 禹山林1, 迟晓元1( ), 张建成1(
), 张建成1( )
)
- 1山东省花生研究所/农业农村部花生生物学与遗传育种重点实验室,266100,山东青岛
2临沂市农业科学院,276012,山东临沂
3德州市临邑县农业农村局,251500,山东德州
Genome-Wide Identification and Characteristic Analyzation of the TCP Transcription Factors Family in Peanut
Wang Tong1( ), Zhao Xiaodong2, Zhen Pingping3, Chen Jing1, Chen Mingna1, Chen Na1, Pan Lijuan1, Wang Mian1, Xu Jing1, Yu Shanlin1, Chi Xiaoyuan1(
), Zhao Xiaodong2, Zhen Pingping3, Chen Jing1, Chen Mingna1, Chen Na1, Pan Lijuan1, Wang Mian1, Xu Jing1, Yu Shanlin1, Chi Xiaoyuan1( ), Zhang Jiancheng1(
), Zhang Jiancheng1( )
)
- 1Shandong Peanut Research Institute/Key Laboratary of Peanut Biology and Genetic Improvement of Ministry of Agriculture and Rural Affairs, Qingdao 266100, Shandong, China
2Linyi Academy of Agricultural Sciences, Linyi 276012, Shandong, China
3Agriculture and Rural Bureau of Linyi County, Dezhou 251500, Shandong, China
摘要:
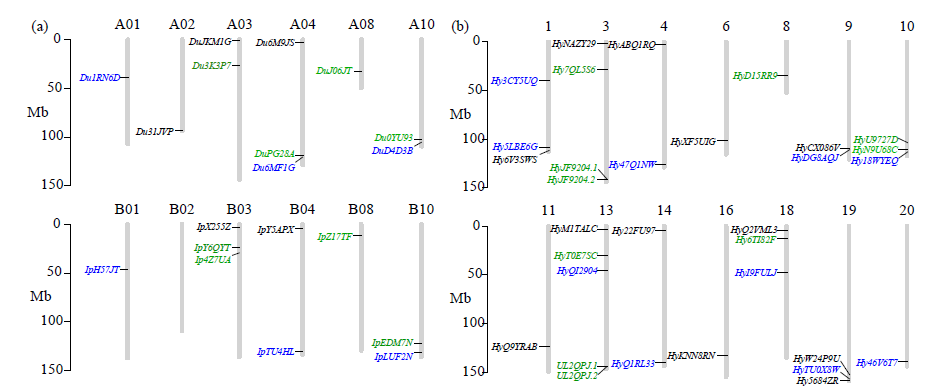
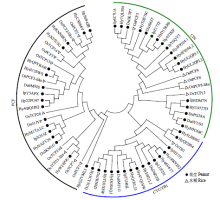
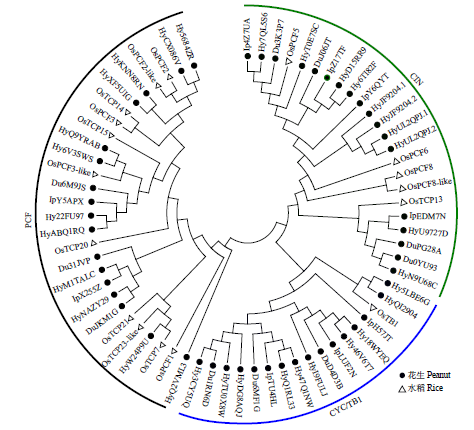
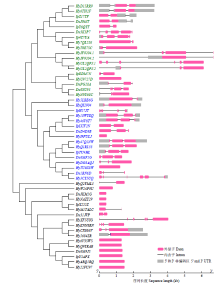
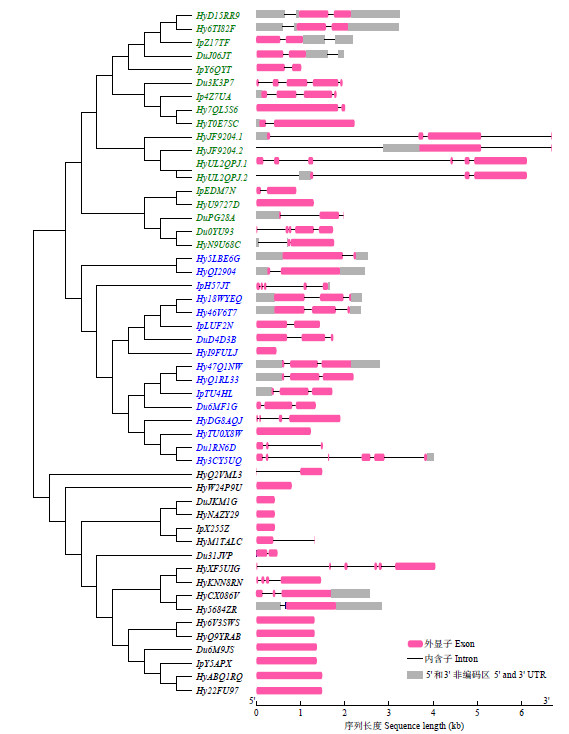
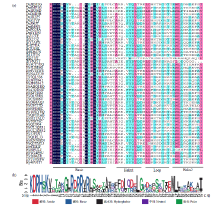
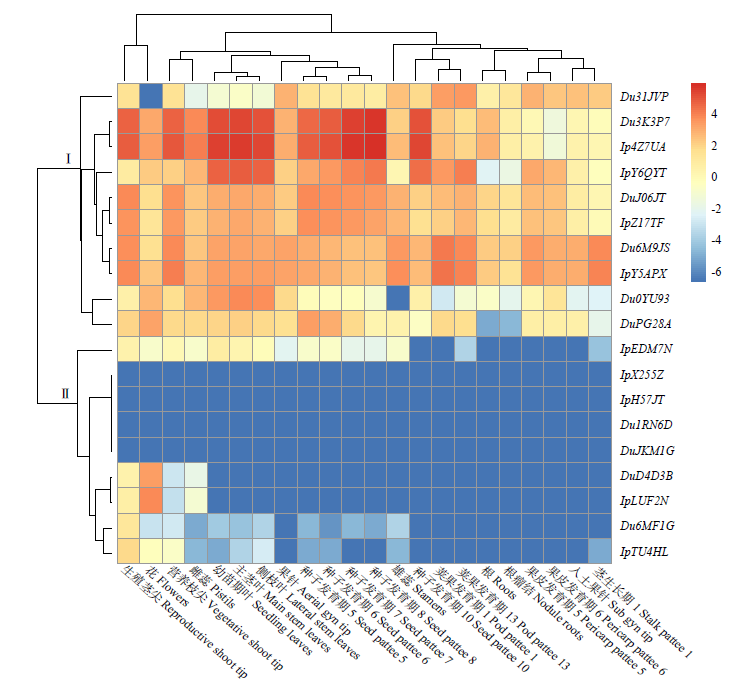
TCP基因家族是植物中一类重要的转录因子,参与植物整个生长发育阶段的调控,尤其在花器官和分生组织中发挥重要作用。目前,在花生中尚无TCP相关基因的报道。为研究花生各TCP转录因子的生物调控作用,以及为进一步分析花生TCP基因提供参考信息,利用生物信息学方法在全基因组水平对花生TCP家族基因进行鉴定,分析其染色体定位、系统进化、基因结构、保守基序和基因表达模式。结果分别从花生野生种和栽培种鉴定出19个和32个TCP基因,不均匀分布在9个野生种染色体和14个栽培种染色体上。系统进化分析表明,51个花生TCP基因可划分为亚家族Ⅰ(PCF)和亚家族Ⅱ两个亚类,其中亚家族Ⅱ包括2个分支,CIN和CYC/TB1。这些基因都含有高度保守的bHLH结构域,但其内含子结构存在较大差异,内含子数量及长度分布与基因的系统进化有较大关系,其中亚家族Ⅰ成员的内含子较少,亚家族Ⅱ内含子较多,且长度差异较大。表达谱分析显示,仅有7个基因在各组织中呈现显著差异性表达,其中有6个基因的差异表达与分生组织和花器官有关,推测其在花生茎尖和花的生长发育过程中起重要作用。
| [1] | Cubas P, Lauter N, Doebley J, et al. The TCP domain:a motif found in proteins regulating plant growth and development. The Plant Journal, 1999,18(2):215-222. |
| [2] | Wang H F, Wang H W, Liu R, et al. Genome-wide identification of TCP family transcription factors in Medicago truncatula reveals significant roles of miR319-targeted TCPs in nodule development. Frontiers in Plant Science, 2018,9:774. |
| [3] | Doebley J, Stec A, Hubbard L. The evolution of apical dominance in maize, Nature, 1997,386(6624):485-488. |
| [4] | Luo D, Carpenter R, Vincent C, et al. Origin of floral asymmetry in Antirrhinum. Nature, 1996,383(6603):794-799. |
| [5] | Aggarwal P, Das Gupta M, Joseph A P. Identification of specific DNA binding residues in the TCP family of transcription factors in Arabidopsis. Plant Cell, 2010,22:1174-1189. |
| [6] | Dhaka N, Bhardwaj V, Sharma M K, et al. Evolving tale of TCPs:new paradigms and old lacunae. Frontiers in Plant Science, 2017,8:479. |
| [7] | Martín-Trillo M, Cubas P. TCP genes:a family snapshot ten years later. Trends in Plant Science, 2010,15(1):31-39. |
| [8] | Zhao J, Zhai Z, Li Y, et al. Genome-wide identification and expression profiling of the TCP family genes in spike and grain development of wheat (Triticum aestivum L.). Frontiers in Plant Science, 2018,9:1282. |
| [9] | Nicolas M, Rodriguezbuey M L, Francozorrilla J M, et al. A recently evolved alternative splice site in the BRANCHED1a gene controls potato plant architecture. Current Biology, 2015,25(14):1799-1809. |
| [10] | Kieffer M, Master V, Waites R, et al. TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. The Plant Journal, 2011,68:147-158. |
| [11] | Nag A, King S, Jack T. MiR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 2009,106(52):22534-22539. |
| [12] | Resentini F, Felipo-Benavent A, Colombo L, et al. TCP14 and TCP15 mediate the promotion of seed germination by gibberellins in Arabidopsis thaliana. Molecular Plant, 2015,8(3):482-485. |
| [13] | Gonzalez-Grandio E, Pajoro A, Franco Zorrilla J M, et al. Abscisic acid signaling is controlled by a BRANCHED1/H D-ZIP I cascade in Arabidopsis axillary buds. Proceedings of the National Academy of Sciences of the United States of America, 2017,114(2):E245-E254. |
| [14] | Zhou M, Li D Y, Li Z G, et al. Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bentgrass. Plant Physiology, 2013,161:1375-1391. |
| [15] | Liu H, Wu M, Li F, et al. TCP Transcription factors in moso bamboo (Phyllostachys edulis):genome-wide identification and expression analysis. Frontiers in Plant Science, 2018,9:1263. |
| [16] | Koyama T, Furutani M, Tasaka M, et al. TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. The Plant Cell, 2007,19(2):473-484. |
| [17] | Takeda T, Amano K, Ohto M A, et al. RNA interference of the Arabidopsis putative transcription factor TCP16 gene results in abortion of early pollen development. Plant Molecular Biology, 2006,61:165-177. |
| [18] | Zheng K, Ni Z, Qu Y, et al. Genome-wide identification and expression analyses of TCP transcription factor genes in Gossypium barbadense. Scientific Reports, 2018,8(1):14526. |
| [19] | Aguilar-Martinez J A, Poza-Carrion C, Cubas P. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell, 2007,19:458-472. |
| [20] | Martin-Trillo M, Grandio E G, Serra F, et al. Role of tomato BRANCHED1- like genes in the control of shoot branching. Plant Journal, 2011,67(4):701-714. |
| [21] | 刘洋, 张慧, 辛大伟, 等. 大豆TCP转录因子家族结构域分析及功能预测. 大豆科学, 2012,31(5):707-713,717. |
| [22] | Parapunova V, Busscher M, Busscher-Lange J, et al. Identification,cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biology, 2014,14(1):157. |
| [23] | Francis A, Dhaka N, Bakshi M, et al. Comparative phylogenomic analysis provides insights into TCP gene functions in Sorghum. Scientific Reports, 2016,6:38488. |
| [24] | Chai W, Jiang P, Huang G, et al. Identification and expression profiling analysis of TCP family genes involved in growth and development in maize. Physiology and Molecular Biology of Plants, 2017,23:779-791. |
| [25] | Leng X, Wei H, Xu X, et al. Genome-wide identification and transcript analysis of TCP transcription factors in grapevine. BMC Genomics, 2019,20(1):786-786. |
| [26] | Bertiolid J, C annon S B, F roenicke L, et al. The genome sequences of Arachis duranensis and Arachis ipaensis,the diploid ancestors of cultivated peanut. Nature Genetics, 2016,48(4):438. |
| [27] | Zhuang W, Chen H, Yang M, et al. The genome of cultivated peanut provides insight into legume karyotypes,polyploid evolution and crop domestication. Nature Genetics, 2019,51(5):865-876. |
| [28] | Xu R, Sun P, Jia F, et al. Genomewide analysis of TCP transcription factor gene family in Malus domestica. Journal of Genetics, 2014,93(3):733-746. |
| [29] | 王亚鹏. 马铃薯StTCP家族基因鉴定及其对块茎休眠解除的响应. 兰州:甘肃农业大学, 2019. |
| [30] | 赵一彤, 魏戏梦, 薛超玲, 等. 枣TCP家族基因鉴定及其表达分析. 河北农业大学学报, 2019,42(5):39-45. |
| [31] | Li W, Li D D, Han L H, et al. Genome-wide identification and characterization of TCP transcription factor genes in upland cotton (Gossypium hirsutum). Scientific Reports, 2017,7(1):10118. |
| [32] | 华方静, 刘风珍, 万勇善, 等. 花生及其野生种质溶血磷脂酸酰基转移酶基因(LPAAT)的克隆及序列分析. 分子植物育种, 2014,12(1):74-79. |
| [33] | Liu Y, Guan X, Liu S, et al. Genome-wide identification and analysis of TCP transcription factors involved in the formation of leafy head in Chinese cabbage. International Journal of Molecular Sciences, 2018,19(3):847. |
| [34] | Huo Y Z, Xiong W D, Su K L, et al. Genome-Wide Analysis of the TCP Gene Family in Switchgrass(Panicum virgatum L.). International Journal of Genomics, 2019(1):1-13. |
| [35] | Feng K, Hao J, Liu J, et al. Genome-wide identification,classification,and expression analysis of TCP transcription factors in carrot. Canadian Journal of Plant Science, 2019,99(4):525-535. |
| [36] | Hubbard L, Mcsteen P, Doebley J, et al. Expression patterns and mutant phenotype of teosinte branched1 correlate with growth suppression in maize and teosinte. Genetics, 2002,162(4):1927-1935. |
| [37] | Finlayson S A. Arabidopsis TEOSINTE BRANCHED1-LIKE 1 regulates axillary bud outgrowth and is homologous to monocot TEOSINTE BRANCHED1. Plant and Cell Physiology, 2007,48(5):667-677. |
| [38] | Luo D, Carpenter R, Copsey L, et al. Control of organ asymmetry in flowers of Antirrhinum. Cell, 1999,99(4):367-376. |
| [39] | Broholm S K, Tähtiharju S, Laitinen R A, et al. A TCP domain transcription factor controls flower type specification along the radial axis of the Gerbera (Asteraceae) inflorescence. Proceedings of the National Academy of Sciences of the United States of America, 2008,105(26):9117-9122. |
| [1] | 修俊杰, 刘学良. 水氮互作对花针期花生生理特性及生长的影响[J]. 作物杂志, 2023, (6): 174–180 |
| [2] | 赵鹏鹏, 李鲁华, 任明见, 安畅, 洪鼎立, 李欣, 徐如宏. 小麦GzCIPK7-5B基因的生物信息学及表达分析[J]. 作物杂志, 2023, (4): 77–84 |
| [3] | 刘素军, 蒙美莲, 苏日古嘎. 干旱胁迫及复水对马铃薯糖代谢途径中基因表达影响的研究[J]. 作物杂志, 2023, (1): 38–45 |
| [4] | 崔亚男, 张曼, 张朋磊, 刘兵, 郝西, 臧秀旺. H2O2对吸胀冷害下花生种子萌发的影响[J]. 作物杂志, 2022, (4): 236–241 |
| [5] | 张小全, 贾振宇, 李举旭, 李洪臣, 王豹祥, 汪健, 石刚, 王川, 武云杰. 不同促根措施对皖南烤烟成熟期钾素代谢的影响[J]. 作物杂志, 2022, (3): 205–210 |
| [6] | 刘菊, 李广存, 段绍光, 胡军, 简银巧, 刘建刚, 金黎平, 徐建飞. 不同夜间温度处理对马铃薯试管薯及块茎形成相关基因表达的影响[J]. 作物杂志, 2022, (3): 92–98 |
| [7] | 赵锴, 晋秀娟, 孙丽丽, 闫荣岳, 卢娟, 郭峰, 史雨刚, 孙黛珍. 小麦脱植基相关基因在春性小麦叶片叶绿素降解过程中的作用分析[J]. 作物杂志, 2022, (2): 81–88 |
| [8] | 刘卫星, 范小玉, 张枫叶, 贺群岭, 陈雷, 李可, 吴继华. 不同前茬和种衣剂用量对花生病虫害及产量的影响[J]. 作物杂志, 2021, (6): 199–204 |
| [9] | 龚林涛, 苏秀娟, 廖燕, 克热木汗·吾斯曼, 周迪, 尹松松. 薰衣草芳樟醇合酶基因的克隆、表达及酶活性检测[J]. 作物杂志, 2021, (6): 78–87 |
| [10] | 蔺儒侠, 郭凤丹, 王兴军, 夏晗, 侯蕾. 花生分子育种研究进展[J]. 作物杂志, 2021, (5): 1–5 |
| [11] | 于天一, 郑亚萍, 邱少芬, 姜大奇, 吴正锋, 郑永美, 孙学武, 沈浦, 王才斌, 张建成. 酸化土壤施钙对不同花生品种(系)钙吸收、利用及产量的影响[J]. 作物杂志, 2021, (4): 80–85 |
| [12] | 索炎炎, 张翔, 司贤宗, 李亮, 余琼, 余辉. 施用磷和钙对花生生长、产量及磷钙利用效率的影响[J]. 作物杂志, 2021, (1): 187–192 |
| [13] | 刘卫星,贺群岭,张枫叶,范小玉,陈雷,李可,吴继华. 大粒花生品种区域试验的AMMI模型分析[J]. 作物杂志, 2020, (2): 60–64 |
| [14] | 权宝全,白冬梅,田跃霞,薛云云. 不同源库关系对花生光合特性及产量的影响[J]. 作物杂志, 2018, (4): 102–105 |
| [15] | 何中国,朱统国,李玉发,王佰众,牛海龙,刘红欣,李伟堂,牟书靓. 吉林省花生育种现状及发展方向[J]. 作物杂志, 2018, (4): 8–12 |
|
||