作物杂志,2023, 第5期: 43–48 doi: 10.16035/j.issn.1001-7283.2023.05.007
利用玉米F1群体进行玉米全株鲜重的全基因组预测分析
- 1吉林农业大学农学院,130118,吉林长春
2定西市农业科学研究院,743000,甘肃定西
3中国农业科学院作物科学研究所,100193,北京
Whole-Genome Predictive Analysis of Fresh Weight per Plant Using the Maize F1 Population
Yang Zongying1( ), Xiao Gui2(
), Xiao Gui2( ), Zhang Hongwei3(
), Zhang Hongwei3( )
)
- 1College of Agronomy, Jilin Agricultural University, Changchun 130118, Jilin, China
2Dingxi Academy of Agricultural Sciences, Dingxi 743000, Gansu, China
3Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100193, China
摘要:
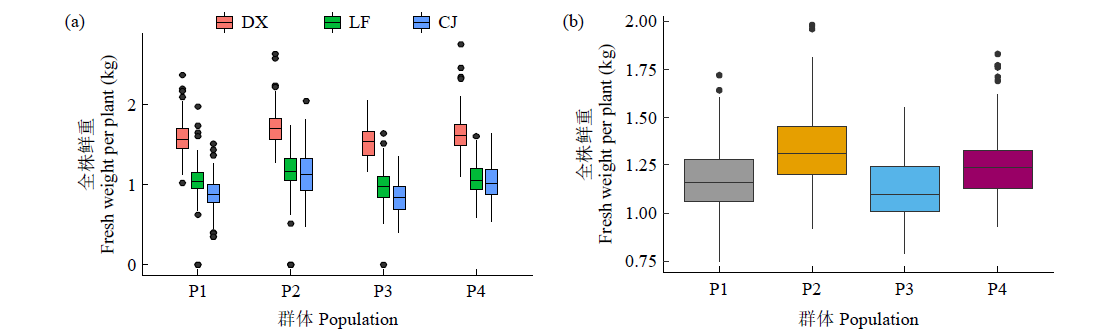
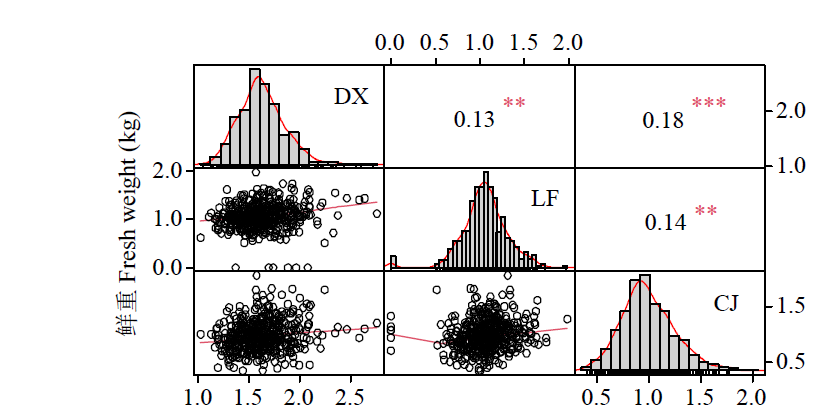
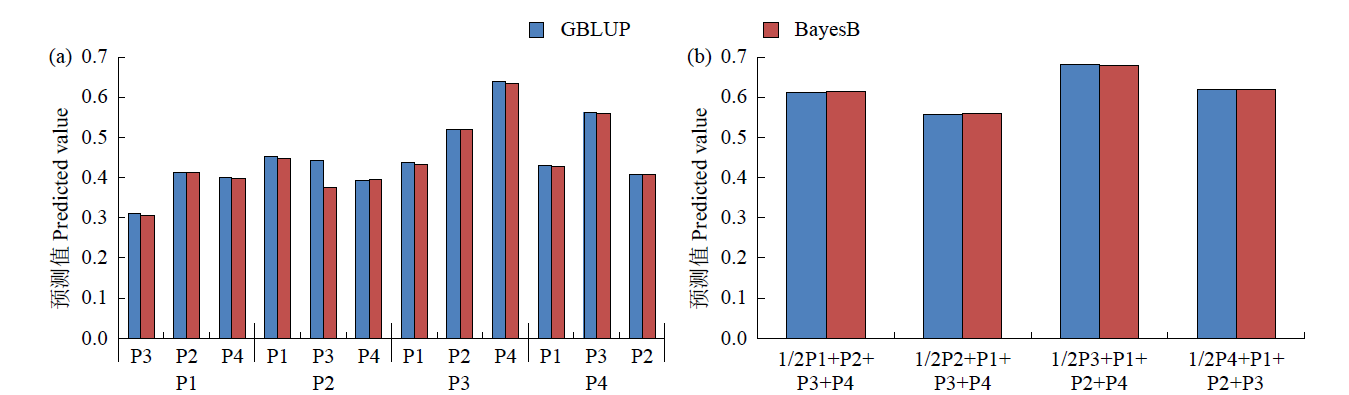
基因组预测是一种可以提高育种效率的分子育种技术。利用中北410(SN915,YH-1)和北农368(60271,2193)的亲本为父本与120个玉米自交系组配4个杂交群体。利用10k的SNP芯片对亲本进行基因分型,在3个环境下对4个群体的全株鲜重进行评价。结果表明,从环境来看,甘肃种植的全株鲜重最高;从群体来看,群体2的鲜重最高;4个群体的变异系数为0.27~0.33,表明4个测交群体的表型变异较大。通过实施一对一(利用其中一个群体为训练群体分别预测其他群体)以及多对一(利用3个群体及第4个群体的一半作为训练群体预测第4个群体的另一半)的预测方案,表明杂交群体间(一对一)的基因组预测准确性低于群体内(多对一)的基因组预测准确性,亲缘关系近的群体间预测效果更好。通过在训练群体中加入与预测群体有亲缘关系的材料可以改进基因组预测的效果。
| [1] |
李少昆, 赵久然, 董树亭, 等. 中国玉米栽培研究进展与展望. 中国农业科学, 2017, 50(11):1941-1959.
doi: 10.3864/j.issn.0578-1752.2017.11.001 |
| [2] | Tilman D, Balzer C, Hill J, et al. Global food demand and the sustainable intensification of agriculture. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108 (50):20260-20264. |
| [3] | 闫亚飞. 河套灌区不同饲草生产性能与品质研究. 北京: 中国农业科学院, 2016. |
| [4] |
Iwata H. Genomic selection in plant breeding: from theory to practice. Briefings in Functional Genomics, 2010, 9(2):166-177.
doi: 10.1093/bfgp/elq001 |
| [5] |
Heffner E L, Lorenz A J, Jannink J L, et al. Plant breeding with genomic selection: gain per unit time and cost. Crop Science, 2010, 50(5):1681-1690.
doi: 10.2135/cropsci2009.11.0662 |
| [6] |
Crossa J, Pérez P, Hickey J M, et al. Genomic prediction in CIMMYT maize and wheat breeding program. Heredity, 2013, 112 (1):48.
doi: 10.1038/hdy.2013.16 |
| [7] |
Li D, Wang P, Gu R, et al. Genetic relatedness and the ratio of subpopulation-common alleles are related in genomic prediction across structured subpopulations in maize. Plant Breeding, 2019, 138(6):802-809.
doi: 10.1111/pbr.v138.6 |
| [8] | Xu S, Zhu D, Zhang Q. Predicting hybrid performance in rice using genomic best linear unbiased prediction. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(34):12456-12461. |
| [9] |
Cui Y, Li R, Li G, et al. Hybrid breeding of rice via genomic selection. Plant Biotechnology Journal, 2020, 18(1):57-67.
doi: 10.1111/pbi.13170 pmid: 31124256 |
| [10] |
Huang X, Yang S, Gong J, et al. Genomic analysis of hybrid rice varieties reveals numerous superior alleles that contribute to heterosis. Nature Communications, 2015, 6(7052):6258.
doi: 10.1038/ncomms7258 |
| [11] | Nickolai A, Tai S, Wang W, et al. SNP-Seek database of SNPs derived from 3000 rice genomes. Nucleic Acids Research, 2015 (D1):1023-1027. |
| [12] | Hadasch S, Simko I, Hayes R J, et al. Comparing the predictive abilities of phenotypic and marker-assisted selection methods in a biparental lettuce population. Plant Genome, 2016, 9(1):1-11. |
| [13] |
Yang J, Mezmouk S, Baumgarten A, et al. Incomplete dominance of deleterious alleles contributes substantially to trait variation and heterosis in maize. PLoS Genetics, 2016, 13(9):e1007019.
doi: 10.1371/journal.pgen.1007019 |
| [14] | Bates D, Mächler M, Bolker B, et al. Fitting linear mixed-effects models using lme4. Statistics & Computing, 2014, 1406(1):133-199. |
| [15] | Hallauer A R, Carena M J, Miranda Filho J B. Quantitative genetics in maize breeding. New York: Springer Science & Business Media, 2010. |
| [16] |
Murray M G, Thompson C L, Wendel J F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research, 1980, 8 (19):4321-4325.
doi: 10.1093/nar/8.19.4321 pmid: 7433111 |
| [17] |
Pérez P, Campos G D L. Genome-wide regression and prediction with the BGLR statistical package. Genetics, 2014, 198(2):483-495.
doi: 10.1534/genetics.114.164442 pmid: 25009151 |
| [18] |
Liu X, Wang H, Wang H, et al. Factors affecting genomic selection revealed by empirical evidence in maize. The Crop Journal, 2018, 6(4):341-352.
doi: 10.1016/j.cj.2018.03.005 |
| [19] |
Dias K O D G, Gezan S A, Guimarães C T, et al. Improving accuracies of genomic predictions for drought tolerance in maize by joint modeling of additive and dominance effects in multi- environment trials. Heredity, 2018, 121(1):24-37.
doi: 10.1038/s41437-018-0053-6 |
| [20] |
Massman J M, Gordillo A, Lorenzana R E, et al. Genomewide predictions from maize single-cross data. Theoretical and Applied Genetics, 2013, 126(1):13-22.
doi: 10.1007/s00122-012-1955-y pmid: 22886355 |
| [21] |
Hayes B J, Bowman P J, Chamberlain A C, et al. Accuracy of genomic breeding values in multi-breed dairy cattle populations. Genetics Selection Evolution, 2009, 41(1):1-9.
doi: 10.1186/1297-9686-41-1 |
| [22] | 刘策, 孟焕文, 程智慧. 植物全基因组选择育种技术原理与研究进展. 分子植物育种, 2020, 18(16):5335-5342. |
| [23] | 陈雨, 姜淑琴, 孙炳蕊, 等. 基因组选择及其在作物育种中的应用. 广东农业科学, 2017, 44(9):1-7. |
| [1] | 曹庆军, 李刚, 杨浩, 娄玉勇, 杨粉团, 孔凡丽, 李辛琲, 赵新凯, 姜晓莉. 不同耕作方式对春玉米种床质量的影响及其与幼苗群体建成和产量的关系[J]. 作物杂志, 2023, (5): 249–254 |
| [2] | 于乐, 李林, 黄红娟, 黄兆峰, 朱文达, 魏守辉. 湖北省玉米田杂草种类组成及群落特征[J]. 作物杂志, 2023, (5): 272–279 |
| [3] | 曲海涛, 李忠南, 王越人, 马艺文, 相洋, 邬生辉, 谭倬, 王纯, 魏强, 罗瑶, 李光发. 玉米百粒重遗传育种效应研究[J]. 作物杂志, 2023, (5): 66–70 |
| [4] | 杨密, 王美娟, 许海涛. 不同生态区玉米自交系苞叶动态发育差异性研究[J]. 作物杂志, 2023, (5): 81–90 |
| [5] | 袁刘正, 王会强, 王秋岭, 朱世蝶, 赵月强, 袁曼曼, 王会涛, 张运栋, 柳家友, 袁永强. 遮阴条件下玉米自交系配合力与遗传效应分析[J]. 作物杂志, 2023, (4): 104–109 |
| [6] | 郑飞, 陈静, 崔亚坤, 孔令杰, 孟庆长, 李杰, 刘瑞响, 张美景, 赵文明, 陈艳萍. 淮北不同生态区丰产稳产宜机收玉米新品种筛选[J]. 作物杂志, 2023, (4): 110–117 |
| [7] | 王丽萍, 白岚方, 王天昊, 王宵璇, 白云鹤, 王玉芬. 不同施氮水平对青贮玉米植株氮素积累和转运的影响[J]. 作物杂志, 2023, (4): 165–173 |
| [8] | 李雨新, 卢敏, 赵久然, 王荣焕, 徐田军, 吕天放, 蔡万涛, 张勇, 薛洪贺, 刘月娥. 京津唐夏玉米区生产现状调研分析[J]. 作物杂志, 2023, (4): 174–181 |
| [9] | 刘松涛, 田再民, 刘子刚, 高志佳, 张静, 贺东刚, 黄智鸿, 兰鑫. 基于转录组测序揭示玉米抗倒伏相关基因和代谢通路[J]. 作物杂志, 2023, (4): 31–37 |
| [10] | 谢文锦, 李方明, 李宁, 杨海龙, 付俊, 张中伟, 高旭东, 丰光. 基于GGE双标图的北方地区鲜食糯玉米产量和品质性状及试点鉴别力分析[J]. 作物杂志, 2023, (4): 85–90 |
| [11] | 温胜慧, 杨俊伟, 王洋, 李公建, 翁建峰, 段灿星, 贾鑫, 王建军. 玉米抗真菌病害基因挖掘与分子育种利用研究进展[J]. 作物杂志, 2023, (3): 1–11 |
| [12] | 常青, 李立军, 渠佳慧, 张艳丽, 韩冬雨, 赵鑫瑶. 土默川平原麦后复种饲用玉米‖油菜模式的增产优势及氮素利用效率[J]. 作物杂志, 2023, (3): 167–174 |
| [13] | 郭书磊, 王瑛, 魏良明, 张新, 刘焱, 吴伟华, 卢道文, 雷晓兵, 王振华, 鲁晓民. 不同生态条件下玉米产量影响因素分析[J]. 作物杂志, 2023, (3): 205–214 |
| [14] | 高沐甜, 邱冠杰, 朱通通, 李瑞莲, 邓敏, 罗红兵, 黄成. 玉米―大刍草渗入系群体的剑叶遗传基础解析[J]. 作物杂志, 2023, (3): 51–57 |
| [15] | 李忠南, 王越人, 马艺文, 相洋, 邬生辉, 曲海涛, 李福林, 张淑琴, 李光发. 基于玉米DH系分离群体叶鞘、花丝、花药与穗轴颜色性状的遗传分析[J]. 作物杂志, 2023, (3): 75–79 |
|
||