作物杂志,2025, 第6期: 100–111 doi: 10.16035/j.issn.1001-7283.2025.06.013
藜麦LEA基因家族鉴定及盐胁迫下的表达分析
张业猛1,2( ), 卢柯欣1, 毛文钰1, 张晓燕1, 李文静1, 杨修1
), 卢柯欣1, 毛文钰1, 张晓燕1, 李文静1, 杨修1
- 1
济宁学院生命科学与工程学院, 273155, 山东曲阜
2中国科学院西北高原生物研究所, 810000, 青海西宁
Identification and Expression Analysis of LEA Gene Family in Chenopodium quinoa Willd. under Salt Stress
Zhang Yemeng1,2( ), Lu Kexin1, Mao Wenyu1, Zhang Xiaoyan1, Li Wenjing1, Yang Xiu1
), Lu Kexin1, Mao Wenyu1, Zhang Xiaoyan1, Li Wenjing1, Yang Xiu1
- 1
School of Life Science and Bioengineering ,Jining University Qufu 273155, Shandong, China
2Northwest Institute of Plateau Biology ,Chinese Academy of Sciences Xining 810000, Qinghai, China
摘要:
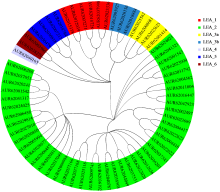
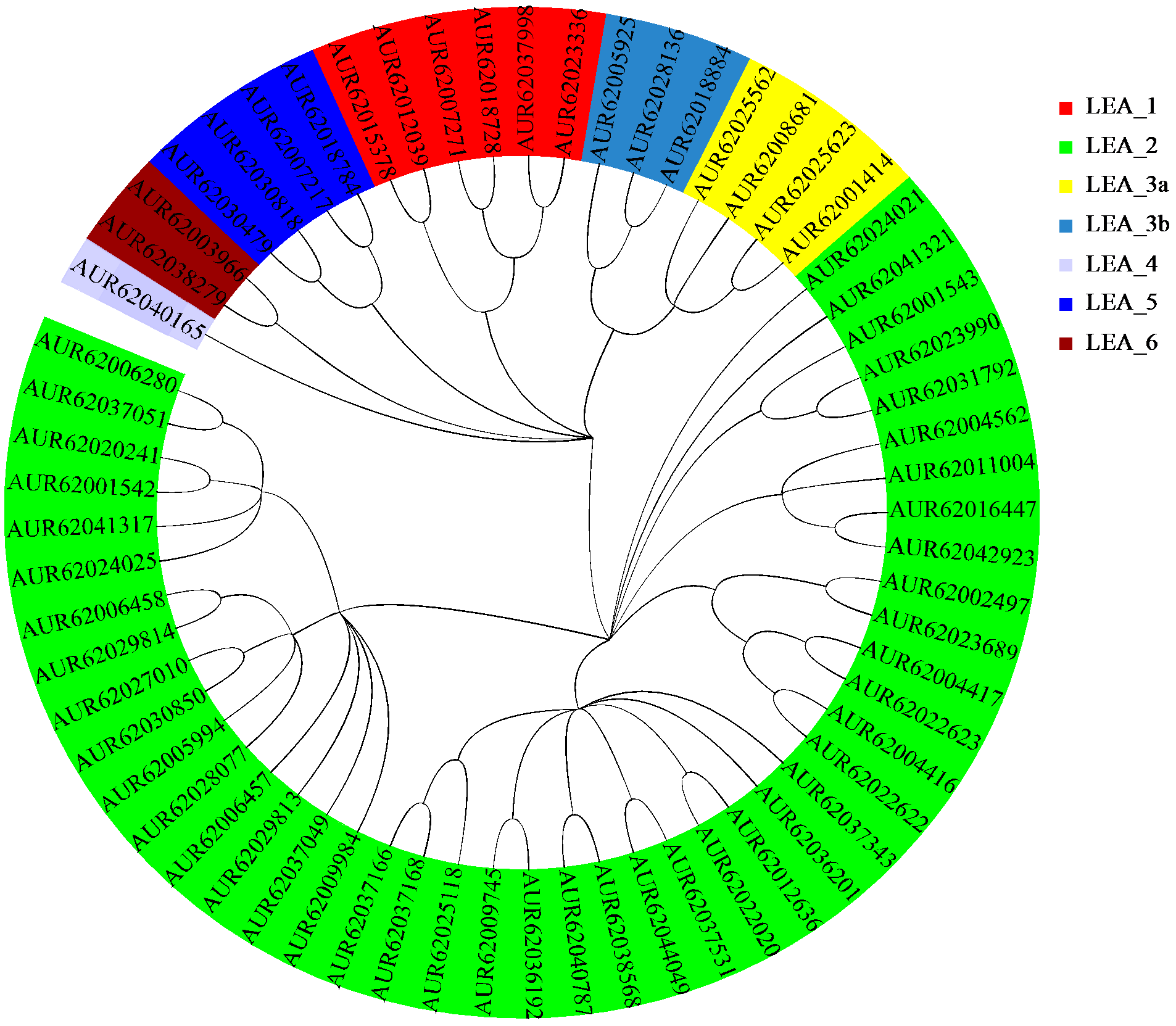
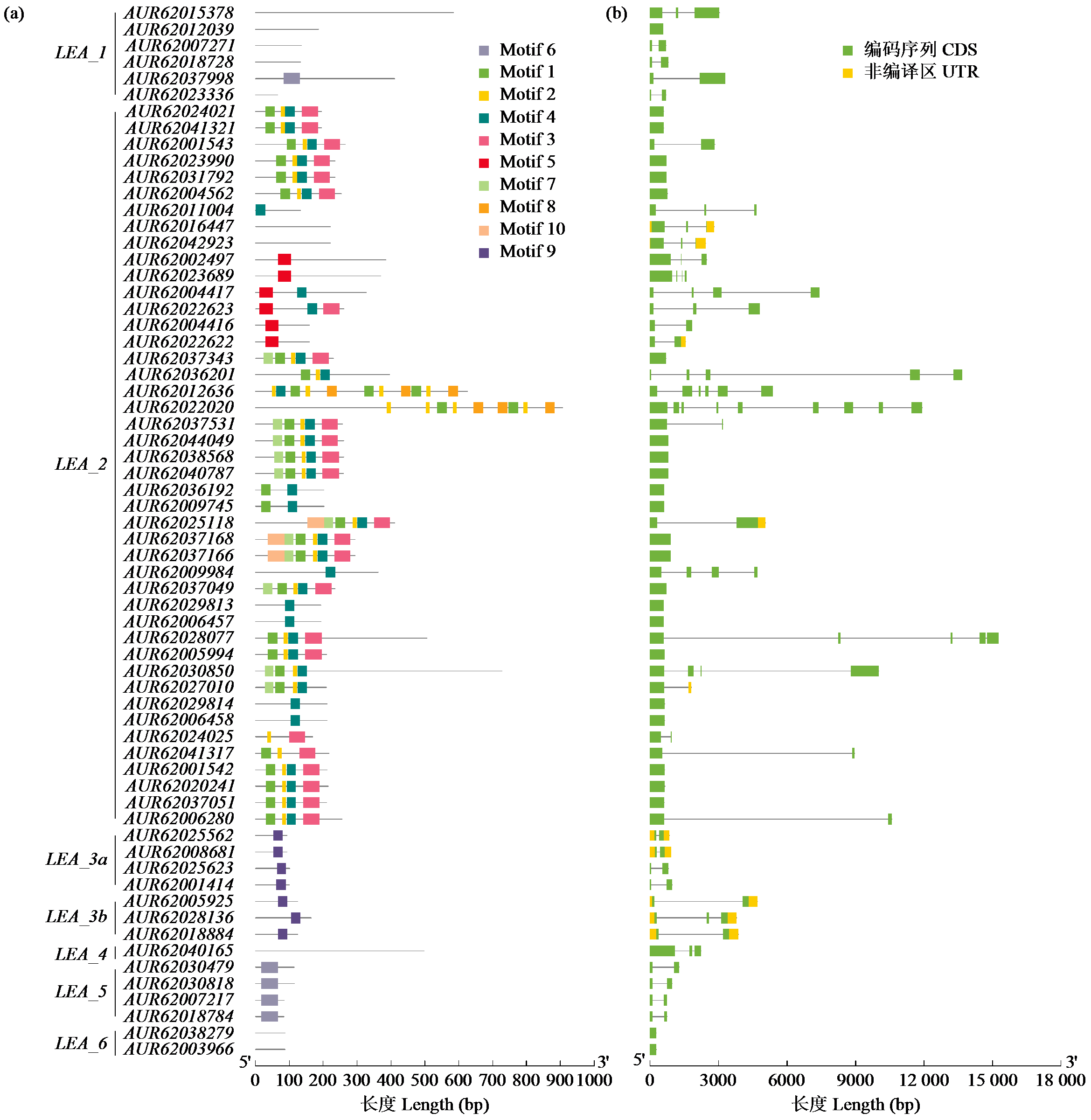
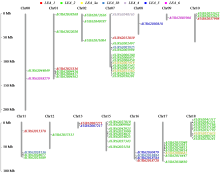
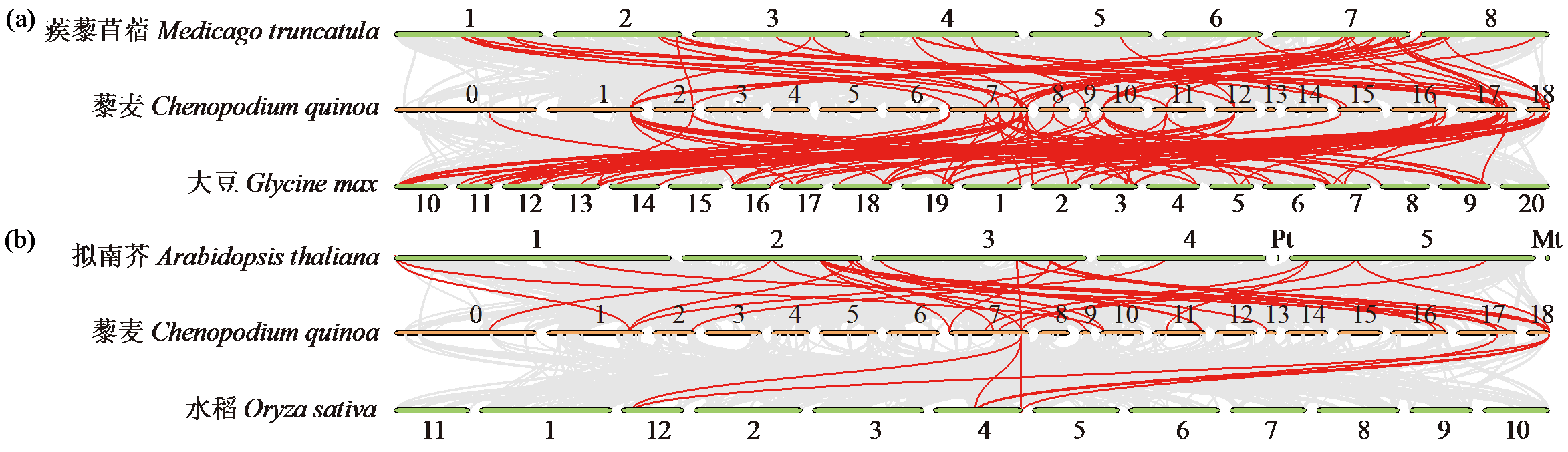
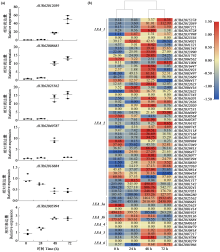
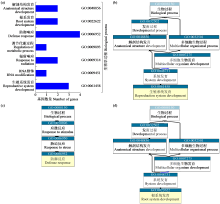
为分析藜麦LEA基因家族的理化性质、染色体分布、基因结构、蛋白保守基序、物种间共线性及表达模式,利用生物信息学方法对藜麦LEAs进行全基因组鉴定。结果显示,在藜麦基因组中共鉴定到64个LEA基因家族成员,分属7个亚组,分布于13条染色体上,同一亚组的成员具有相似的基因结构和保守基序。种内共线性分析发现,64个LEA基因存在20个重复基因对;种间共线性分析发现,其与豆科植物之间具有高度的进化保守性。LEA基因的启动子区域存在多种激素和非生物胁迫相关的作用元件,其中46个LEA基因受盐胁迫诱导表达,通过GO注释发现主要参与解剖结构发育、根系发育、防御响应和生殖系统发育等生物学过程,各亚组基因发挥不同作用以提升藜麦盐胁迫耐受性。
| [1] |
Miyazaki K, Ohkubo Y, Yasui H, et al. Overexpression of rice OsLEA5 relieves the deterioration in seed quality caused by high- temperature stress. Plant Biotechnology, 2021, 38(3):367-371.
doi: 10.5511/plantbiotechnology.21.0603a pmid: 34782824 |
| [2] |
Vidović M, Battisti I, Pantelić A, et al. Desiccation tolerance in Ramonda serbica panc.: an integrative transcriptomic, proteomic, metabolite and photosynthetic study. Plants, 2022, 11(9):1199.
doi: 10.3390/plants11091199 |
| [3] |
Dure L Ⅲ, Greenway S C, Galau G A. Developmental biochemistry of cottonseed embryogenesis and germination: changing messenger ribonucleic acid populations as shown by in vitro and in vivo protein synthesis. Biochemistry, 1981, 20(14):4162-4168.
doi: 10.1021/bi00517a033 pmid: 7284317 |
| [4] |
Huang R L, Xiao D, Wang X, et al. Genome-wide identification,evolutionary and expression analyses of LEA gene family in peanut (Arachis hypogaea L.). BMC Plant Biology, 2022, 22(1):155.
doi: 10.1186/s12870-022-03462-7 |
| [5] |
Jia C P, Guo B, Wang B K, et al. The LEA gene family in tomato and its wild relatives: genome-wide identification, structural characterization, expression profiling, and role of SlLEA6 in drought stress. BMC Plant Biology, 2022, 22(1):596.
doi: 10.1186/s12870-022-03953-7 |
| [6] |
Singh K K, Graether S P. The in vitro structure and functions of the disordered late embryogenesis abundant three proteins. Protein Science, 2021, 30(3):678-692.
doi: 10.1002/pro.4028 pmid: 33474748 |
| [7] | Furuki T, Sakurai M. Group 3 LEA protein model peptides protect enzymes against desiccation stress. Acta Biochimica et Biophysica Sinica, 2016, 1864(9):1237-1243. |
| [8] |
Nadarajah K K. ROS homeostasis in abiotic stress tolerance in plants. International Journal of Molecular Sciences. 2020, 21(15):5208.
doi: 10.3390/ijms21155208 |
| [9] |
Zhou C C, Niu S H, El-Kassaby Y A, et al. Genome-wide identification of late embryogenesis abundant protein family and their key regulatory network in Pinus tabuliformis cold acclimation. Tree Physiology, 2023, 43(11):1964-1985.
doi: 10.1093/treephys/tpad095 |
| [10] |
Yang Z F, Mu Y H, Wang Y Q, et al. Characterization of a novel TtLEA2 gene from Tritipyrum and its transformation in wheat to enhance salt tolerance. Frontiers in Plant Science, 2022, 13:830848.
doi: 10.3389/fpls.2022.830848 |
| [11] |
Cheng Z H, Zhang X M., Yao W J. et al. Genome-wide search and structural and functional analyses for late embryogenesis- abundant (LEA) gene family in poplar. BMC Plant Biology, 2021, 21(1):110.
doi: 10.1186/s12870-021-02872-3 |
| [12] |
Hinojosa L, González J, Barrios-Masias F, et al. Quinoa abiotic stress responses: a review. Plants, 2018, 7(4):106.
doi: 10.3390/plants7040106 |
| [13] |
Younis I Y, Sedeek M S, Essa A F, et al. Exploring geographic variations in quinoa grains: unveiling anti-alzheimer activity via GC-MS, LC-QTOF-MS/MS, molecular networking, and chemometric analysis. Food Chemistry, 2025, 465:141918.
doi: 10.1016/j.foodchem.2024.141918 |
| [14] |
Jarvis D E, Ho Y S, Lightfoot D J, et al. The genome of Chenopodium quinoa. Nature, 2017, 542:307-312.
doi: 10.1038/nature21370 |
| [15] |
Zhao Y Y, Hao Y P, Dong Z Y, et al. Identification and expression analysis of LEA gene family members in pepper (Capsicum annuum L.). FEBS Open Bio, 2023, 13(12):2246-2262.
doi: 10.1002/feb4.v13.12 |
| [16] | 贾冰晨, 王宇, 张东亮, 等. 藜麦内参基因筛选及盐胁迫相关基因表达分析. 烟台大学学报(自然科学与工程版), 2020, 33(3):283-288. |
| [17] |
Zhao X X, Wang S Y, Guo F G, et al. Genome-wide identification of polyamine metabolism and ethylene synthesis genes in Chenopodium quinoa Willd. and their responses to low-temperature stress. BMC Genomics, 2024, 25(1):370.
doi: 10.1186/s12864-024-10265-7 |
| [18] |
Wang Q, Lei X J, Wang Y H, et al. Genome-wide identification of the LEA gene family in Panax ginseng: evidence for the role of PgLEA2-50 in plant abiotic stress response. Plant Physiology and Biochemistry, 2024, 212:108742.
doi: 10.1016/j.plaphy.2024.108742 |
| [19] |
Chen Y K, Li C H, Zhang B, et al. The role of the late embryogenesis-abundant (LEA) protein family in development and the abiotic stress response: a comprehensive expression analysis of potato (Solanum tuberosum). Genes, 2019, 10(2):148.
doi: 10.3390/genes10020148 |
| [20] |
Li Y F, Xu Y H, Ma Z W. Comparative analysis of the exon- intron structure in eukaryotic genomes. Yangtze Medicine, 2017, 1(1):50-64.
doi: 10.4236/ym.2017.11006 |
| [21] |
Hibshman J D, Goldstein B. LEA motifs promote desiccation tolerance in vivo. BMC Biology, 2021, 19(1):263.
doi: 10.1186/s12915-021-01176-0 pmid: 34903234 |
| [22] |
Chen Y, Shen J, Zhang L, et al. Nuclear translocation of OsMFT 1 that is impeded by OsFTIP1 promotes drought tolerance in rice. Molecular Plant, 2021, 14(8):1297-1311.
doi: 10.1016/j.molp.2021.05.001 |
| [23] |
Lu Y, Sun J, Yang Z M, et al. Genome-wide identification and expression analysis of glycine-rich RNA-binding protein family in sweet potato wild relative Ipomoea trifida. Gene, 2019, 686:177-186.
doi: S0378-1119(18)31187-9 pmid: 30453066 |
| [24] |
Hu M Q, Li Z Q, Lin X J, et al. Comparative analysis of the LEA gene family in seven Ipomoea species, focuses on sweet potato (Ipomoea batatas L.). BMC Plant Biology, 2024, 24:1256.
doi: 10.1186/s12870-024-05981-x |
| [25] |
Jia J S, Ge N, Wang Q Y, et al. Genome-wide identification and characterization of members of the LEA gene family in Panax notoginseng and their transcriptional responses to dehydration of recalcitrant seeds. BMC Genomics, 2023, 24(1):126.
doi: 10.1186/s12864-023-09229-0 |
| [26] |
Zhang Y J, Fan N N, Wen W W, et al. Genome-wide identification and analysis of LEA_2 gene family in alfalfa (Medicago sativa L.) under aluminum stress. Frontiers in Plant Science, 2022, 13:976160.
doi: 10.3389/fpls.2022.976160 |
| [27] | Liu S Y, Zhang L, Sang Y P, et al. Demographic history and natural selection shape patterns of deleterious mutation load and barriers to introgression across Populus genome. Molecular Biology and Evolution, 2022, 39(2):msac008. |
| [1] | 陈奕, 陈潇, 张茂星, 李静, 常静静, 李嘉炜, 林海晴, 陈兴平, 邓晓亮, 谢大森, 郭少龙, 沈仲灯, 张白鸽. 不同冬瓜品种对低盐胁迫的响应及其耐盐性综合评价[J]. 作物杂志, 2025, (6): 58–66 |
| [2] | 滕文, 叶凡, 周舟, 王屿乐, 刘立军. 小麦和油菜秸秆还田处理对盐胁迫下水稻产量和品质的影响[J]. 作物杂志, 2025, (5): 11–18 |
| [3] | 闫晶蓉, 庞春花, 张永清, 毋悦悦, 侯钰晨, 王嘉祺, 乔曼. 脱硫石膏与腐植酸配施对盐碱地土壤及藜麦生长的影响[J]. 作物杂志, 2025, (5): 47–53 |
| [4] | 王兖薇, 武俊喜, 汪艳, 牟涛, 朗卓玛, 苗彦军. 盐碱胁迫对异株荨麻种子萌发的影响[J]. 作物杂志, 2025, (5): 67–73 |
| [5] | 李小雨, 黄杰, 杨钊, 柴继宽, 杨发荣, 魏玉明, 刘文瑜, 拜伟俊. 基于AHP法的观赏藜麦综合评价体系的建立与应用[J]. 作物杂志, 2025, (3): 70–77 |
| [6] | 张圣昌, 魏玉明, 马丽娜, 杨钊, 刘文瑜, 黄杰, 刘欢, 杨发荣. 种植密度和施肥对饲用型藜麦生长特性的影响[J]. 作物杂志, 2025, (2): 128–134 |
| [7] | 景茂雅, 张子玉, 张萌, 合佳敏, 严翻翻, 高艳梅, 张永清. 水杨酸浸种对盐胁迫藜麦种子萌发及幼苗生长的影响[J]. 作物杂志, 2025, (1): 194–201 |
| [8] | 李峰, 高宏云, 张翀, 张宝英, 马建富, 郭娜, 白苇, 方爱国, 杨志敏, 李源. 盐胁迫对燕麦生长及生理指标的影响[J]. 作物杂志, 2024, (6): 140–146 |
| [9] | 鄂利锋, 徐金崇, 陈修斌, 权建华, 华军, 尹丽娟, 王舜奇, 赵文勤. 外源硅对盐胁迫下娃娃菜种子萌发及幼苗生理特性的影响[J]. 作物杂志, 2024, (6): 212–217 |
| [10] | 马丽娜, 魏玉明, 文莉芳, 张学俭, 杨钊, 黄杰, 张圣昌, 李小雨, 刘欢, 杨发荣. 云南元谋地区22份藜麦种质的农艺性状及营养品质分析[J]. 作物杂志, 2024, (6): 47–54 |
| [11] | 侯钰晨, 庞春花, 张永清, 康书瑜, 毋悦悦, 闫晶蓉, 王嘉祺. 施用生物炭与氮肥对盐碱胁迫下藜麦幼苗生理生长特性的影响[J]. 作物杂志, 2024, (4): 240–246 |
| [12] | 顾怀应, 胡诗钦, 赵晴, 刘长华, 孟丽君. 根际微生物增强水稻耐盐性研究进展[J]. 作物杂志, 2024, (4): 8–13 |
| [13] | 刘建霞, 王文庆, 薛乃雯, 郭绪虎, 马赛雅, 朱国芳, 温日宇. 14个不同产地藜麦种质染色体核型分析[J]. 作物杂志, 2024, (3): 82–89 |
| [14] | 合佳敏, 张永清, 张萌, 梁萍, 王丹, 严翻翻. 烯效唑浸种对盐碱胁迫下藜麦农艺性状及生理特性的影响[J]. 作物杂志, 2024, (2): 234–241 |
| [15] | 吕宝莲, 杨宇昕, 崔立操, 史峰, 马亮, 孔秀英, 张立超, 倪志勇. 小麦bHLH家族转录因子的鉴定及其在盐胁迫条件下的表达分析[J]. 作物杂志, 2024, (1): 65–72 |
|
||