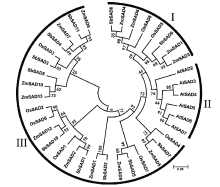
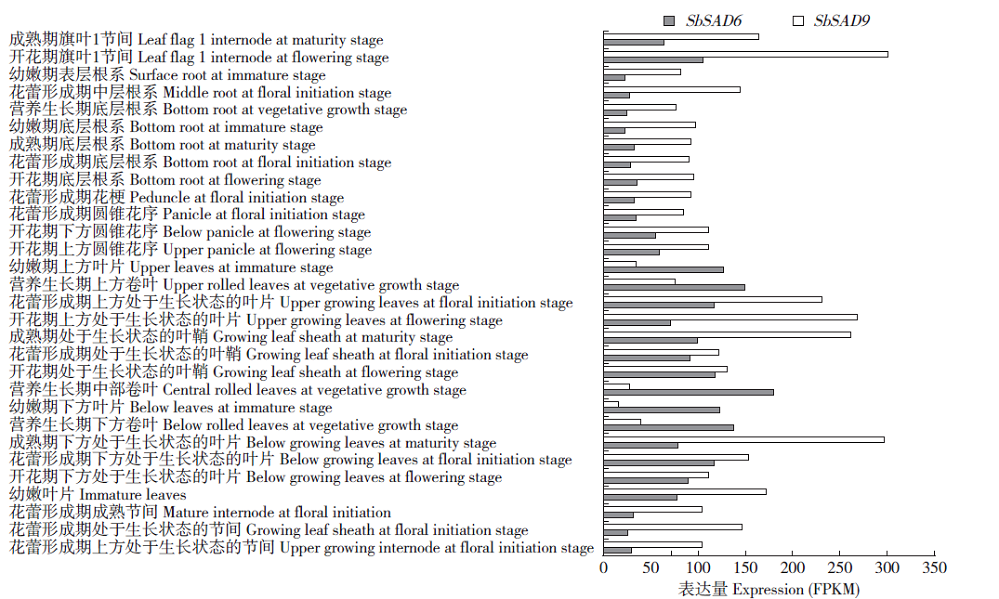
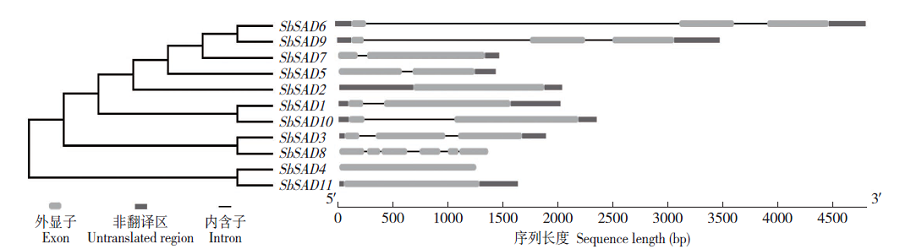
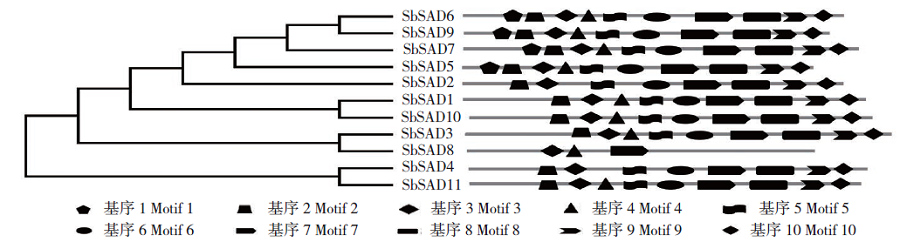
Crops ›› 2020, Vol. 36 ›› Issue (2): 20-27.doi: 10.16035/j.issn.1001-7283.2020.02.004
Previous Articles Next Articles
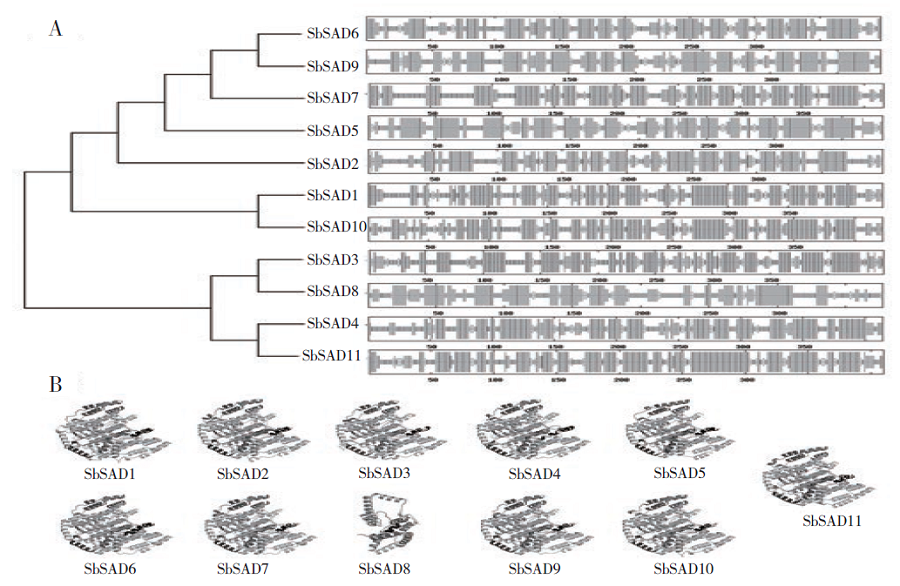
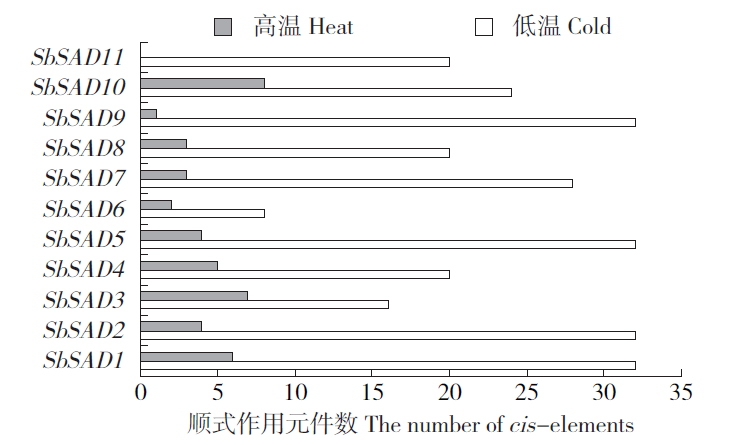
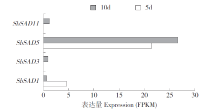
Identification of Stearyl -ACP Desaturase Gene (SbSAD) Family and Their Expression Analysis at Different Developmental Stages in Sorghum
Zhao Xunchao,Xu Jingyu( ),Gai Shengnan,Wei Yulei,Xu Xiaoxuan,Ding Dong,Liu Meng,Zhang Jinjie,Shao Wenjing
),Gai Shengnan,Wei Yulei,Xu Xiaoxuan,Ding Dong,Liu Meng,Zhang Jinjie,Shao Wenjing
- College of Agronomy, Heilongjiang Bayi Agricultural University/Key Laboratory of Modern Agricultural Cultivation and Crop Germplasm Improvement of Heilongjiang Province, Daqing 163319, Heilongjiang, China
| [1] | 杨士春 . 甜高粱籽粒中脂肪酸含量的气相色谱分析. 中国粮油学报, 2012,27(11):100-104. |
| [2] | 张国琴, 葛玉彬, 张正英 , 等. 高粱抗旱研究综述. 甘肃农业科技, 2018,56(6):67-72. |
| [3] | O'Byrne D J, Knauft D A, Shireman R B . Low fat-monounsaturated rich diets containing high-oleic peanuts improve serum lipoprotein profiles. Lipids, 1997,32(7):687-695. |
| [4] | Carrillo C, Del M C M, Roelofs H , et al. Activation of human neutrophils by oleic acid involves the production of reactive oxygen species and a rise in cytosolic calcium concentration:a comparison with N-6 polyunsaturated fatty acids. Cellular Physiology and Biochemistry, 2011,28(2):329-338. |
| [5] | 袁蕊, 敖宗华, 丁海龙 , 等. 高粱中脂肪酸和低分子有机酸气相色谱测定. 酿酒, 2011,38(4):42-43. |
| [6] | Ohlrogge J, Browse J . Lipid biosynthesis. The Plant Cell, 1995,7(7):957-970. |
| [7] | Xuan W Y, Zhang Y, Liu Z Q , et al. Molecular cloning and expression analysis of a novel BCCP subunit gene from Aleurites moluccana. Genetics and Molecular Research, 2015,14(3):9922-9931. |
| [8] | Gonzalez-Thuillier I, Venegas-Caleron M, Sanchez R , et al. Sunflower (Helianthus annuus) fatty acid synthase complex:beta-hydroxyacyl-[acyl carrier protein] dehydratase genes. Planta, 2016,243(2):397-410. |
| [9] | Rodriguez M F, Sanchez-Garcia A, Salas J J , et al. Characterization of soluble acyl-ACP desaturases from Camelina sativa,Macadamia tetraphylla and Dolichandra unguiscati. Journal of Plant Physiology, 2015,178(15):35-42. |
| [10] | Li-Beisson Y, Shorrosh B, Beisson F , et al. Acyl-lipid metabolism. The Arabidopsis Book, 2013,11:e0161. |
| [11] | Kachroo A, Shanklin J, Whittle E , et al. The Arabidopsis stearoyl-acyl carrier protein-desaturase family and the contribution of leaf isoforms to oleic acid synthesis. Plant Molecular Biology, 2007,63(2):257-271. |
| [12] | Ruddle P N, Whetten R, Cardinal A , et al. Effect of a novel mutation in a △9-stearoyl-ACP-desaturase on soybean seed oil composition. Theoretical and Applied Genetics, 2013,126(1):241-249. |
| [13] | Jung S, Tate P L, Horn R , et al. The phylogenetic relationship of possible progenitors of the cultivated peanut. Journal of Heredity, 2003,94(4):334-340. |
| [14] | Knutzon D S, Thompson G A, Radke S E , et al. Modification of Brassica seed oil by antisense expression of a stearoyl-acyl carrier protein desaturase gene. Proceedings of the National Academy of Sciences of the United States of America, 1992,89(7):2624-2628. |
| [15] | Slocombe S P, Cummins I, Jarvis R P , et al. Nucleotide sequence and temporal regulation of a seed-specific Brassica napus cDNA encoding a stearoyl-acyl carrier protein (ACP) desaturase. Plant Molecular Biology, 1992,20(1):151-155. |
| [16] | Shanklin J, Somerville C . Stearoyl-acyl-carrier-protein desaturase from higher plants is structurally unrelated to the animal and fungal homologs. Proceedings of the National Academy of Sciences of the United States of America, 1991,88(6):2510-2514. |
| [17] | McKeon T A, Stumpf P K . Purification and characterization of the stearoyl-acyl carrier protein desaturase and the acyl-acyl carrier protein thioesterase from maturing seeds of safflower. The Journal of Biological Chemistry, 1982,257(20):12141-12147. |
| [18] | Zhang Y, Maximova S N, Guiltinan M J . Characterization of a stearoyl-acyl carrier protein desaturase gene family from chocolate tree,Theobroma cacao L. Frontiers in Plant Science, 2015,6:239-251. |
| [19] | Lightner J, Wu J, Browse J . A mutant of Arabidopsis with increased levels of stearic acid. Plant Physiology, 1994,106(4):1443-1451. |
| [20] | Osorio J, Fernándezmartínez J, Mancha M , et al. Mutant sunflowers with high concentration of saturated fatty acids in the oil. Crop Science, 1995,35(3):739-742. |
| [21] | Craig W, Lenzi P, Scotti N , et al. Transplastomic tobacco plants expressing a fatty acid desaturase gene exhibit altered fatty acid profiles and improved cold tolerance. Transgenic Research, 2008,17(5):769-782. |
| [22] | Byfield G E, Xue H, Upchurch R G . Two genes from soybean encoding soluble Δ9 stearoyl-ACP desaturases. Crop Science, 2006,46(2):840-846. |
| [23] | Murata N, Ishizaki-Nishizawa O, Higashi S , et al. Genetically engineered alteration in the chilling sensitivity of plants. Nature, 1992,356:710-713. |
| [24] | Kodama H, Horiguchi G, Nishiuchi T , et al. Fatty acid desaturation during chilling acclimation is one of the factors involved in conferring low-temperature tolerance to young tobacco leaves. Plant Physiology, 1995,107(4):1177-1185. |
| [25] | Orlova I V, Serebriiskaya T S, Popov V , et al. Transformation of tobacco with a gene for the thermophilic acyl-lipid desaturase enhances the chilling tolerance of plants. Plant and Cell Physiology, 2003,44(4):447-450. |
| [26] | Dong C G, Cao N, Zhang Z G , et al. Characterization of the fatty acid desaturase genes in cucumber:structure,phylogeny,and expression patterns. PLoS ONE, 2016,11(3):e0149917. |
| [27] | Tocher D R, Leaver M J, Hodgson P A . Recent advances in the biochemistry and molecular biology of fatty acyl desaturases. Progress in Lipid Research, 1998,37(2/3):73-117. |
| [28] | Han Y, Xu G, Du H , et al. Natural variations in stearoyl-acp desaturase genes affect the conversion of stearic to oleic acid in maize kernel. Theoretical and Applied Genetics, 2017,130(1):151-161. |
| [29] | Shang X, Cheng C, Ding J , et al. Identification of candidate genes from the SAD gene family in cotton for determination of cottonseed oil composition. Molecular Genetics and Genomics, 2017,292(1):173-186. |
| [30] | Merlo A O, Cowen N, Delate T , et al. Ribozymes targeted to stearoyl-ACP △9 desaturase mRNA produce heritable increases of stearic acid in transgenic maize leaves. Plant Cell, 1998,10:1603-1621. |
| [1] | Gao Jie,Li Qingfeng,Li Xiaorong,Feng Guangcai,Peng Qiu. Analysis of the Characteristics of Dry Matter Production and Light Energy Utilization of Waxy Sorghum Applied in Different Eras in Guizhou Province [J]. Crops, 2020, 36(1): 41-46. |
| [2] | Fan Xinqi,Wang Haiyan,Nie Meng′en,Zhao Xingkui,Zhang Yizhong,Yang Huiyong,Zhang Xiaojuan,Liang Du,Duan Yonghong,Liu Qingshan. Effects of EMS Mutagenesis on Emergence and Agronomic Traits in Sorghum [J]. Crops, 2020, 36(1): 47-54. |
| [3] | Song Jian,Cao Xiaoning,Wang Haigang,Chen Ling,Wang Junjie,Liu Sichen,Qiao Zhijun. Identification and Expression Analysis of ASR Family Genes in Setaria italica [J]. Crops, 2019, 35(6): 33-42. |
| [4] | Tang Taoxia,Wang Zhihe,Shi Zhiguo,Chang Ying,Zhang Yingying,Li Yanrong. Research on the Absorption of Heavy Metals by Different Genotypes in Sweet Sorghum [J]. Crops, 2019, 35(6): 50-56. |
| [5] | Liang Xiaohong,Zhang Ruidong,Huang Minjia,Liu Jing,Cao Xiong. Interaction of Film Mulching and Nitrogen Application on Yield, Water and Nitrogen Use Efficiency of Sorghum [J]. Crops, 2019, 35(5): 135-142. |
| [6] | Wang Jinsong,Dong Erwei,Jiao Xiaoyan,Wu Ailian,Bai Wenbin,Wang Lige,Guo Jun,Han Xiong,Liu Qingshan. Effects of Different Planting Patterns on Yield and Nutrient Absorption of Sorghum Jinnuo 3 [J]. Crops, 2019, 35(5): 166-172. |
| [7] | Yue Linqi,Shi Weiping,Guo Jiahui,Guo Pingyi,Guo Jie. Response of Cutin Synthetic Genes of Foxtail Millet to Drought Stress [J]. Crops, 2019, 35(4): 183-190. |
| [8] | Gao Jie,Li Qingfeng,Li Xiaorong,Feng Guangcai,Peng Qiu. Variation Analysis of Agronomic Traits of Waxy Sorghum Varieties (Lines) in Different Eras in Guizhou Province [J]. Crops, 2019, 35(4): 17-23. |
| [9] | Yang Junkai,Shen Yang,Cai Xiaoxi,Wu Shengyang,Li Jianwei,Sun Mingzhe,Jia Bowei,Sun Xiaoli. Genome-Wide Identification and Expression Patterns Analysis of the PHD Family Protein in Glycine max [J]. Crops, 2019, 35(3): 55-65. |
| [10] | Li Chunhong,Lu Xianglong,Zhang Peitong,Su Yanjing,Wang Yiming,Guo Wenqi,Yin Jianmei,Han Xiaoyong,Wang Li,Huo Enjie. Screening Herbicides to Control Weeds for Sweet Sorghum [J]. Crops, 2018, 34(6): 158-161. |
| [11] | Lü Liangjie,Chen Xiyong,Zhang Yelun,Liu Qian,Wang Limei,Ma Le,Li Hui. Bioinformatics Identification of GASA Gene Family Expression Profiles in Wheat [J]. Crops, 2018, 34(6): 58-67. |
| [12] | Zhang Yizhong,Zhou Fuping,Zhang Xiaojuan,Shao Qiang,Yang Bin,Liu Qingshan. Identification and Cluster Analysis of Photosynthetic Characters and WUE in Sorghum Germplasm [J]. Crops, 2018, 34(5): 45-53. |
| [13] | Zhang Ruidong,Cao Xiong,Yue Zhongxiao,Liang Xiaohong,Liu Jing,Huang Minjia. Effects of Nitrogen and Density Interaction on Grain Yield and Nitrogen Use Efficiency of Sorghum [J]. Crops, 2018, 34(5): 110-115. |
| [14] | Zhang Jianhua,Guo Ruifeng,Cao Changlin,Fan Na,Jiang Baiyang,Li Guang,Shi Lijuan,Peng Zhidong,Bai Wenbin. Study on Effect and Safety of Controlling Weed in Sorghum Field by Several Stem and Leaf Treatment Herbicide [J]. Crops, 2018, 34(5): 162-166. |
| [15] | Jie Gao,Qingfeng Li,Qiu Peng,Xiaoyan Jiao,Jinsong Wang. Effects of Different Nutrient Combinations on Plant Production and Nitrogen, Phosphorus and Potassium Utilization Characteristics in Waxy Sorghum [J]. Crops, 2018, 34(4): 138-142. |
|