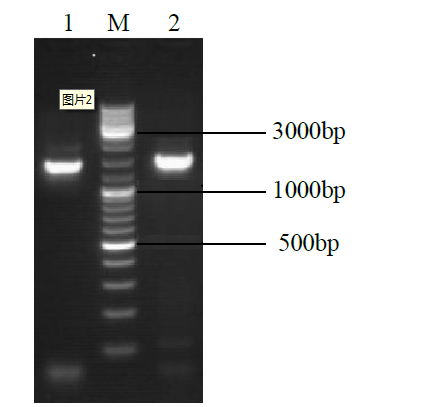
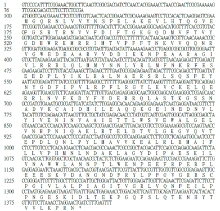
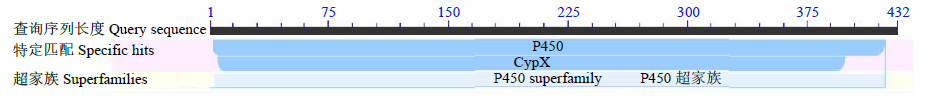
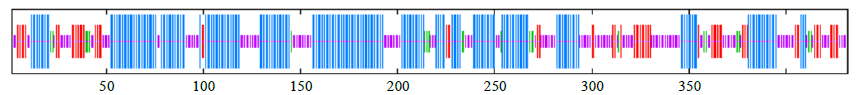
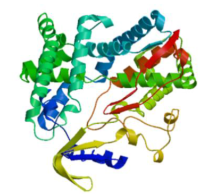
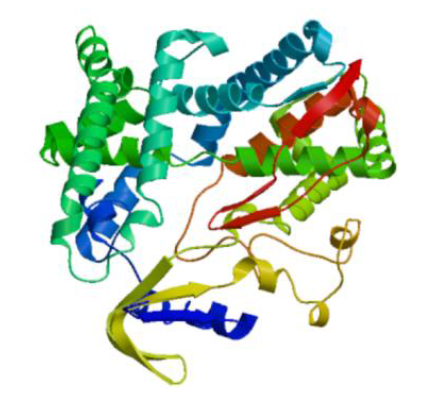
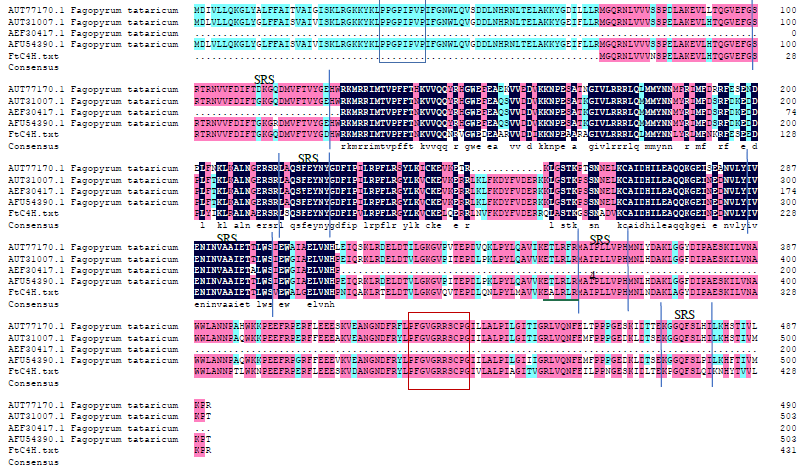
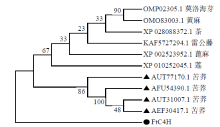
Crops ›› 2022, Vol. 38 ›› Issue (1): 77-83.doi: 10.16035/j.issn.1001-7283.2022.01.011
Previous Articles Next Articles
Cloning and Bioinformatics Analysis of FtC4H Gene from Tartary Buckwheat
Yin Guifang1( ), Duan Ying2, Yang Xiaolin2, Cai Suyun2, Wang Yanqing1, Lu Wenjie1, Sun Daowang1, He Runli2(
), Duan Ying2, Yang Xiaolin2, Cai Suyun2, Wang Yanqing1, Lu Wenjie1, Sun Daowang1, He Runli2( ), Wang Lihua1(
), Wang Lihua1( )
)
- 1Biotechnology and Germplasm Resources Institute, Yunnan Academy of Agricultural Sciences/ Yunnan Provincial Key Laboratory of Agricultural Biotechnology/Key Laboratory of Southwestern Crop Gene Resources and Germplasm Innovation, Ministry of Agriculture and Rural Affairs, Kunming 650205, Yunnan, China
2College of Traditional Chinese Medicine and Food Engineering, Shanxi University of Chinese Medicine, Taiyuan 030619, Shanxi, China
| [1] | 中国植物志编辑委员会. 中国植物志. 第25卷. 北京: 科学出版社, 1998. |
| [2] | 阮池银. 云南小凉山彝族苦荞文化的环境人类学研究. 昆明:云南大学, 2012. |
| [3] |
Bai C Z, Feng M L, Hao X L, et al. Rutin,quercetin,and free amino acid analysis in buckwheat (Fagopyrum) seeds from different locations. Genetics and Molecular Research, 2015, 14(4):19040-19048.
doi: 10.4238/2015.December.29.11 pmid: 26782554 |
| [4] |
Hu Y, Hou Z, Yi R, et al. Tartary buckwheat flavonoids ameliorate high fructose-induced insulin resistance and oxidative stress associated with the insulin signaling and Nrf2/HO-1 pathways in mice. Food and Function, 2017, 8(8):2803-2816.
doi: 10.1039/C7FO00359E |
| [5] |
Bao T, Wang Y, Li Y T, et al. Antioxidant and antidiabetic properties of tartary buckwheat rice flavonoids after in vitro digestion. Journal of Zhejiang University-Science B, 2016, 17(12):941-951.
doi: 10.1631/jzus.B1600243 |
| [6] |
Liu C L, Chen Y S, Yang J H, et al. Antioxidant activity of tartary (Fagopyrum tataricum (L.) Gaertn.) and common (Fagopyrum esculentum Moench) buckwheat sprouts. Journal of Agricultural and Food Chemistry, 2008, 56(1):173-178.
doi: 10.1021/jf072347s |
| [7] | 李玉英, 赵淑娟, 白崇智, 等. 苦荞异槲皮苷对人胃癌细胞SGC-7901增殖及凋亡的影响. 食品科学, 2014, 35(3):193-197. |
| [8] |
Hou Z X, Hu Y Y, Yang X B, et al. Antihypertensive effects of tartary buckwheat flavonoids by improvement of vascular insulin sensitivity in spontaneously hypertensive rats. Food and Function, 2017: 8(11):4217-4228.
doi: 10.1039/C7FO00975E |
| [9] |
Choi S Y, Choi J Y, Lee J M, et al. Tartary buckwheat on nitric oxide-induced inflammation in RAW264.7 macrophage cells. Food and Function, 2015, 6(8):2664-2670.
doi: 10.1039/C5FO00639B |
| [10] |
Russell D W, Conn E E. The cinnamic acid 4-hydroxylase of pea seedlings. Archives of Biochemistry and Biophysics, 1967, 122(1):256-258.
pmid: 4383827 |
| [11] |
Schilmiller A L, Stout J, Weng J K, et al. Mutations in the cinnamate 4-hydroxylase gene impact metabolism,growth and development in Arabidopsis. The Plant Journal, 2009, 60(5):771-782.
doi: 10.1007/s10725-019-00494-2 |
| [12] |
Park N I, Park J H, Park S U. Overexpression of cinnamate 4-hydroxylase gene enhances biosynthesis of decursinol angelate in Angelica gigas hairy roots. Molecular Biotechnology, 2012, 50(2):114-120.
doi: 10.1007/s12033-011-9420-8 pmid: 21626264 |
| [13] |
Singh K, Kumar S, Rani A, et al. Phenylalanine ammonia-lyase (PAL) and cinnamate 4-hydroxylase (C4H) and catechins (flavan-3-ols) accumulation in tea. Functional and Integrative Genomics, 2009, 9(1):125-134.
doi: 10.1007/s10142-008-0092-9 |
| [14] | 曾祥玲, 郑日如, 罗靖, 等. 桂花C4H基因的克隆与表达特性分析. 园艺学报, 2016, 43(3):525-537. |
| [15] | 程俊, 程曦, 盛玲玲, 等. 砀山酥梨肉桂酸4-羟化酶基因的克隆及表达分析. 农业生物技术学报, 2016, 24(11):1698-1708. |
| [16] |
Millar D J, Long M, Donovan G, et al. Introduction of sense constructs of cinnamate 4-hydroxylase (CYP73A24) in transgenic tomato plants shows opposite effects on flux into stem lignin and fruit flavonoids. Phytochemistry, 2007, 68(11):1497-1509.
pmid: 17509629 |
| [17] | Baek M H, Chung B Y, Kim J H, et al. cDNA cloning and expression pattern of Cinnamate-4-Hydroxylase in the Korean blackraspberry. Biochemistry and Molecular Biology Reports, 2008, 41(7):529-536. |
| [10] | Liu W, Zhu D, Liu D, et al. Comparative metabolic activity related to flavonoid synthesis in leaves and flowers of Chrysanthemum morifoliumin response to K deficiency. Plant and Soil, 2010, 335,325-337. |
| [18] |
Cheng S Y, Yan J P, Meng X X, et al. Characterization and expression patterns of a cinnamate-4-hydroxylase gene involved in lignin biosynthesis and in response to various stresses and hormonal treatments in Ginkgo biloba. Acta Physiologiae Plantarum, 2018, 40:1-15.
doi: 10.1007/s11738-017-2577-4 |
| [19] | 黄利娜, 吴光斌, 匡凤元, 等. 莲雾果实C4H基因的克隆及在NO处理下的表达分析. 集美大学学报(自然科学版), 2020, 25(2):105-112. |
| [20] | 陈鸿翰, 袁梦求, 李双江, 等. 苦荞肉桂酸羟化酶基因(FtC4H)的克隆及其UV-B胁迫下的组织表达. 农业生物技术学报, 2013, 21(2):137-147. |
| [21] | 刘荣华, 王丽, 孙朝霞, 等. 苦荞FtC4H基因的cDNA克隆及生物信息学分析. 山西农业大学学报(自然科学版), 2017, 37(11):767-773. |
| [22] | 王轶男, 陈雪, 盖颖. 毛白杨木质素合成酶基因F5H克隆与生物信息学分析. 广东农业科学, 2014, 41(20):131-135. |
| [23] |
Fahrendorf T, Dixon R A. Stress responses in alfalfa (Medicago sativa L.). XVIII:Molecular cloning and expression of the elicitor-inducible cinnamic acid 4-hydroxylase cytochrome P450. Archives of Biochemistry and Biophysics, 1993, 305:509-515.
pmid: 8373188 |
| [24] | Teutsch H G, Hasenfratz M P, Lesot A, et al. Isolation and sequence of a cDNA encoding the Jerusalem artichoke cinnamate 4-hydroxylase,a major plant cytochrome P450 involved in the general phenylpropanoid pathway. Proceedings of the National Academy of Sciences of the United States of America, 1993, 90(9):4102-4106. |
| [11] | Yan Q, Si J, Cui X X, et al. The soybean cinnamate 4-hydroxylase gene GmC4H1 contributes positively to plant defense via increasing lignin content. Plant Growth Regulation, 2019, 88(2):139-149. |
| [25] | 冯艺川, 赵洋, 全雪丽, 等. 膜荚黄芪C4H基因的克隆及表达分析. 分子植物育种, 2021, 19(1):130-136. |
| [1] | Weng Wenfeng, Wu Xiaofang, Zhang Kaixuan, Tang Yu, Jiang Yan, Ruan Jingjun, Zhou Meiliang. The Overexpression of FtbZIP5 Improves Accumulation of Flavonoid in the Hairy Roots of Tartary Buckwheat and Its Salt Tolerance [J]. Crops, 2021, 37(4): 1-9. |
| [2] | Jia Ruiling, Zhao Xiaoqin, Nan Ming, Chen Fu, Liu Yanming, Wei Liping, Liu Junxiu, Ma Ning. Genetic Diversity Analysis and Comprehensive Assessment of Agronomic Traits of 64 Tartary Buckwheat Germplasms [J]. Crops, 2021, 37(3): 19-27. |
| [3] | Jin Jiangang, Tian Zaifang. Grey Correlation Analysis of Introduced Tartary Buckwheat in the Northern Shanxi [J]. Crops, 2021, 37(2): 52-56. |
| [4] | Ma Mingchuan, Liu Longlong, Liu Zhang, Zhou Jianping, Nan Chenghu, Zhang Lijun. Analysis of SSR Loci in Whole Genome and Development of Molecular Markers in Tartary Buckwheat [J]. Crops, 2021, 37(1): 38-46. |
| [5] | Lu Xiaoling, He Ming, Zhang Kaixuan, Liao Zhiyong, Zhou Meiliang. Study on the Cloning and Transformation of Rhamnose Transferase FtF3GT1 Gene in Tartary Buckwheat [J]. Crops, 2020, 36(5): 33-40. |
| [6] | Yang Xuele, Zhang Lu, Li Zhiqing, He Luqiu. Diversity Analysis of Tartary Buckwheat Germplasms Based on Phenotypic Traits [J]. Crops, 2020, 36(5): 53-58. |
| [7] | Li Chunhua, Huang Jinliang, Yin Guifang, Wang Yanqing, Lu Wenjie, Sun Daowang, Wang Chunlong, Guo Laichun, Hong Bo, Ren Changzhong, Wang Lihua. Genetic Analysis of Grain Shape Related Traits in Tartary Buckwheat [J]. Crops, 2020, 36(3): 42-46. |
| [8] | Chengrui Ma,Dabing Xiang,Yan Wan,Jianyong Ouyang,Yue Song,Zhengsong Tang,Jianying Liu,Gang Zhao. Difference Analysis of Spatial Distribution Characteristics of Different Tartary Buckwheat Varieties [J]. Crops, 2020, 36(1): 35-40. |
| [9] | Yang Tian,Zhang Yongqing,Dong Fuhui,Ma Xingxing,Xue Xiaojiao. Research on the Root Growth of Different Drought-Resistant Fagopyrum tataricum under Different Water Conditions [J]. Crops, 2019, 35(6): 76-82. |
| [10] | Yue Linqi,Shi Weiping,Guo Jiahui,Guo Pingyi,Guo Jie. Response of Cutin Synthetic Genes of Foxtail Millet to Drought Stress [J]. Crops, 2019, 35(4): 183-190. |
| [11] | Song Lifang,Feng Meichen,Zhang Meijun,Xiao Lujie,Wang Chao,Yang Wude,Song Xiaoyan. Effects of Exogenous Selenium on the Growth and Development of Tartary Buckwheat and Selenium Content in Grains [J]. Crops, 2019, 35(3): 150-154. |
| [12] | Ma Mingchuan,Liu Longlong,Zhang Lijun,Cui Lin,Zhou Jianping. Morphological Identification and Analysis of EMS-Induced Mutants from Ciqiao [J]. Crops, 2019, 35(3): 37-41. |
| [13] | Yasong Cui, Yan Wang, Lijuan Yang, Chaoxin Wu, Piao Zhou, Pan Ran, Qingfu Chen. Genetic Analysis of Fruit Hull Rate and Related Traits on Tartary Buckwheat [J]. Crops, 2019, 35(2): 51-60. |
| [14] | Haibin Luo, Shengli Jiang, Chengmei Huang, Huiqing Cao, Zhinian Deng, Kaichao Wu, Lin Xu, Zhen Lu, Yuanwen Wei. Cloning and Expression of ScHAK10 Gene in Sugarcane [J]. Crops, 2018, 34(4): 53-61. |
| [15] | Yu Fan,Hongli Wang,Feng He,Dili Lai,Jiajun Wang,Yue Song,Dabing Xiang. Nutritional Quality in Seeds of Tartary Buckwheat Affected by After-Ripening [J]. Crops, 2018, 34(1): 96-101. |
|
||