| [1] |
唐宇, 邵继荣, 周美亮. 中国荞麦属植物分类学的修订. 植物遗传资源学报, 2019,20(3):646-653.
|
| [2] |
Liu Y X, Cai C Z, Yao Y L, et al. Alteration of phenolic profiles and antioxidant capacities of common buckwheat and tartary buckwheat produced in China upon thermal processing. Journal of the Science of Food and Agriculture, 2019,99(12):5565-5576.
|
| [3] |
Kanter M, Aktas C, Erboga M. Protective effects of quercetin against apoptosis and oxidative stress in streptozotocin-induced diabetic rat testis. Food and Chemical Toxicology, 2012,50(3/4):719-725.
|
| [4] |
Wahid A, Ghazanfar A. Possible involvement of some secondary metabolites in salt tolerance of sugarcane. Journal of Plant Physiology, 2006,163(7):723-730.
|
| [5] |
Annamaria G, Marco M, Moira M, et al. Effect of nitrogen starvation on the phenolic metabolism and antioxidant properties of yarrow (Achillea collina Becker ex Rchb.). Food Chemistry, 2008,114(1):204-211.
|
| [6] |
Ariel F D, Manavella P A, Dezar C A, et al. The true story of the HD-Zip family. Trends in Plant Science, 2007,12(9):419-426.
|
| [7] |
Liu M, Wen Y, Sun W, et al. Genome-wide identification,phylogeny,evolutionary expansion and expression analyses of bZIP transcription factor family in tartaty buckwheat. BMC Genomics, 2019,20(1):483.
|
| [8] |
Guo X L, Hou X M, Fang J, et al. The rice GERMINATION DEFECTIVE 1,encoding a B3 domain transcriptional repressor,regulates seed germination and seedling development by integrating GA and carbohydrate metabolism. The Plant Journal, 2013,75(3):403-416.
|
| [9] |
Corrêa L G, Riaño-Pachón D M, Schrago C G, et al. The role of bZIP transcription factors in green plant evolution:adaptive features emerging from four founder genes. PLoS ONE, 2008,3(8):e2944.
|
| [10] |
Li X Y, Gao S Q, Tang Y M, et al. Genome-wide identification and evolutionary analyses of bZIP transcription factors in wheat and its relatives and expression profiles of anther development related TabZIP genes. BMC Genomics, 2015,16:976.
|
| [11] |
Liu J X, Srivastava R, Che P, et al. Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. The Plant Journal, 2007,51(5):897-909.
|
| [12] |
Zhang S X, Haider I, Kohlen W, et al. Function of the HD-Zip I gene Oshox22 in ABA-mediated drought and salt tolerances in rice. Plant Molecular Biology, 2012,80(6):571-585.
|
| [13] |
Lakra N, Nutan K K, Das P, et al. A nuclear-localized histone-gene binding protein from rice (OsHBP1b) functions in salinity and drought stress tolerance by maintaining chlorophyll content and improving the antioxidant machinery. Journal of Plant Physiology, 2015,176:36-46.
|
| [14] |
Hartmann U, Sagasser M, Mehrtens F, et al. Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB,BZIP,and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Molecular Biology, 2005,57(2):155-171.
|
| [15] |
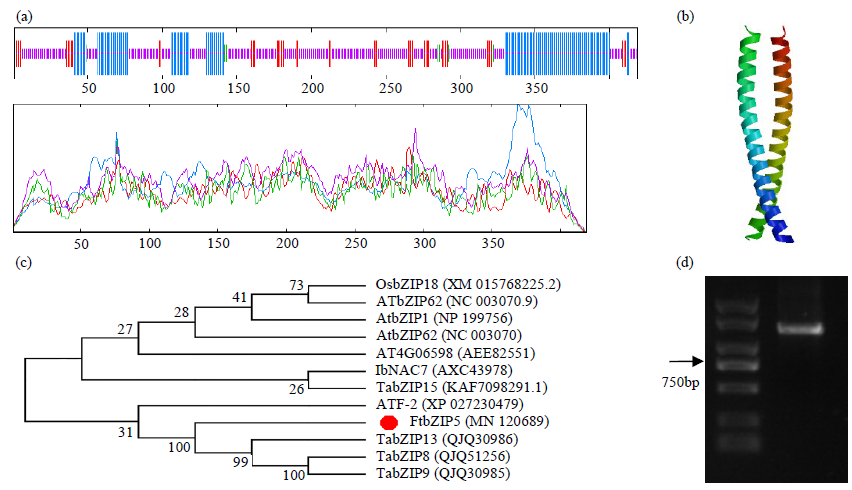
Li Q, Zhao H X, Wang X L, et al. Tartary buckwheat transcription factor FtbZIP5,regulated by FtSnRK2.6,can improve salt/drought resistance in transgenic Arabidopsis. International Journal of Molecular Sciences, 2020,21(3):1123.
|
| [16] |
Malacarne G, Coller E, Czemmel S, et al. The grapevine VvibZIPC22 transcription factor is involved in the regulation of flavonoid biosynthesis. Journal of Experimental Botany, 2016,67(11):3509-3522.
|
| [17] |
卢晓玲, 何铭, 张凯旋, 等. 苦荞鼠李糖基转移酶FtF3GT1基因的克隆与转化毛状根研究. 作物杂志, 2020(5):33-40.
|
| [18] |
谈天斌, 卢晓玲, 张凯旋, 等. TrMYB308基因的克隆及在苦荞毛状根中的功能分析. 植物遗传资源学报, 2019,20(6):1542-1553.
|
| [19] |
李为喜, 朱志华, 李国营, 等. AlCl3分光光度法测定荞麦种质资源中黄酮的研究. 植物遗传资源学报, 2008,9(4):502-505.
|
| [20] |
Park N I, Li X H, Uddin R M, et al. Phenolic compound production by different morphological phenotypes in hairy root cultures of Fagopyrum tataricum Gaertn. Archives of Biological Sciences, 2011,63(1):193-198.
|
| [21] |
苏文华, 张光飞, 李秀华, 等. 植物药材次生代谢产物的积累与环境的关系. 中草药, 2005(9):139-142.
|
| [22] |
许盼云, 吴玉霞, 何天明. 植物对盐碱胁迫的适应机理研究进展. 中国野生植物资源, 2020,39(10):41-49.
|
| [23] |
Zhan X, Shen Q, Chen J, et al. Rice sulfoquinovosyl transferase SQD2.1 mediates flavonoid glycosylation and enhances tolerance to osmotic stress. Plant Cell Environment, 2019,42(7):2215-2230.
|
| [24] |
Dong N Q, Sun Y, Guo T, et al. UDP-glucosyltransferase regulates grain size and abiotic stress tolerance associated with metabolic flux redirection in rice. Nature Communications, 2020,11(1):2629.
|
| [25] |
Pedranzani H, Sierra-de-Grado R, Vigliocco A, et al. Cold and water stresses produce changes in endogenous jasmonates in two populations of Pinus pinaster Ait. Plant Growth Regulation, 2007,52:111-116.
|
| [26] |
Li X H, Kim Y B, Kim Y, et al. Differential stress-response expression of two flavonol synthase genes and accumulation of flavonols in tartary buckwheat. Journal of Plant Physiology, 2013,170(18):1630-1636.
|
| [27] |
Abdallah S B, Aung B, Amyot L, et al. Salt stress (NaCl) affects plant growth and branch pathways of carotenoid and flavonoid biosyntheses in Solanum nigrum. Acta Physiologiae Plantarum, 2016,38(3):72.
|
| [28] |
王琰, 陈建文, 狄晓艳. 水分胁迫下不同油松种源SOD、POD、MDA及可溶性蛋白比较研究. 生态环境学报, 2011,20(10):1449-1453.
|
| [29] |
刘会超, 贾文庆. 盐胁迫对白三叶茎的POD、CAT的影响研究. 吉林农业科学, 2009,34(1):43-46.
|
 ), Wu Xiaofang2, Zhang Kaixuan2, Tang Yu3, Jiang Yan1, Ruan Jingjun1(
), Wu Xiaofang2, Zhang Kaixuan2, Tang Yu3, Jiang Yan1, Ruan Jingjun1( ), Zhou Meiliang2(
), Zhou Meiliang2( )
)