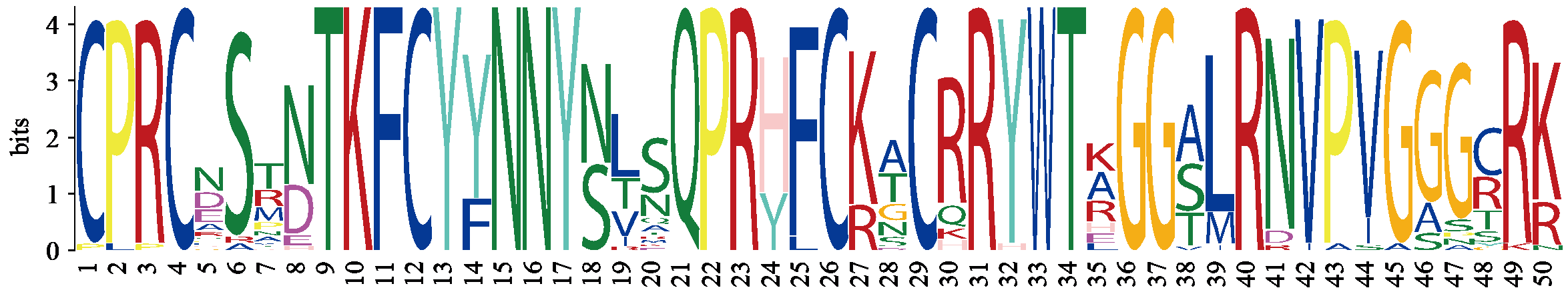Crops ›› 2025, Vol. 41 ›› Issue (1): 54-65.doi: 10.16035/j.issn.1001-7283.2025.01.007
Previous Articles Next Articles
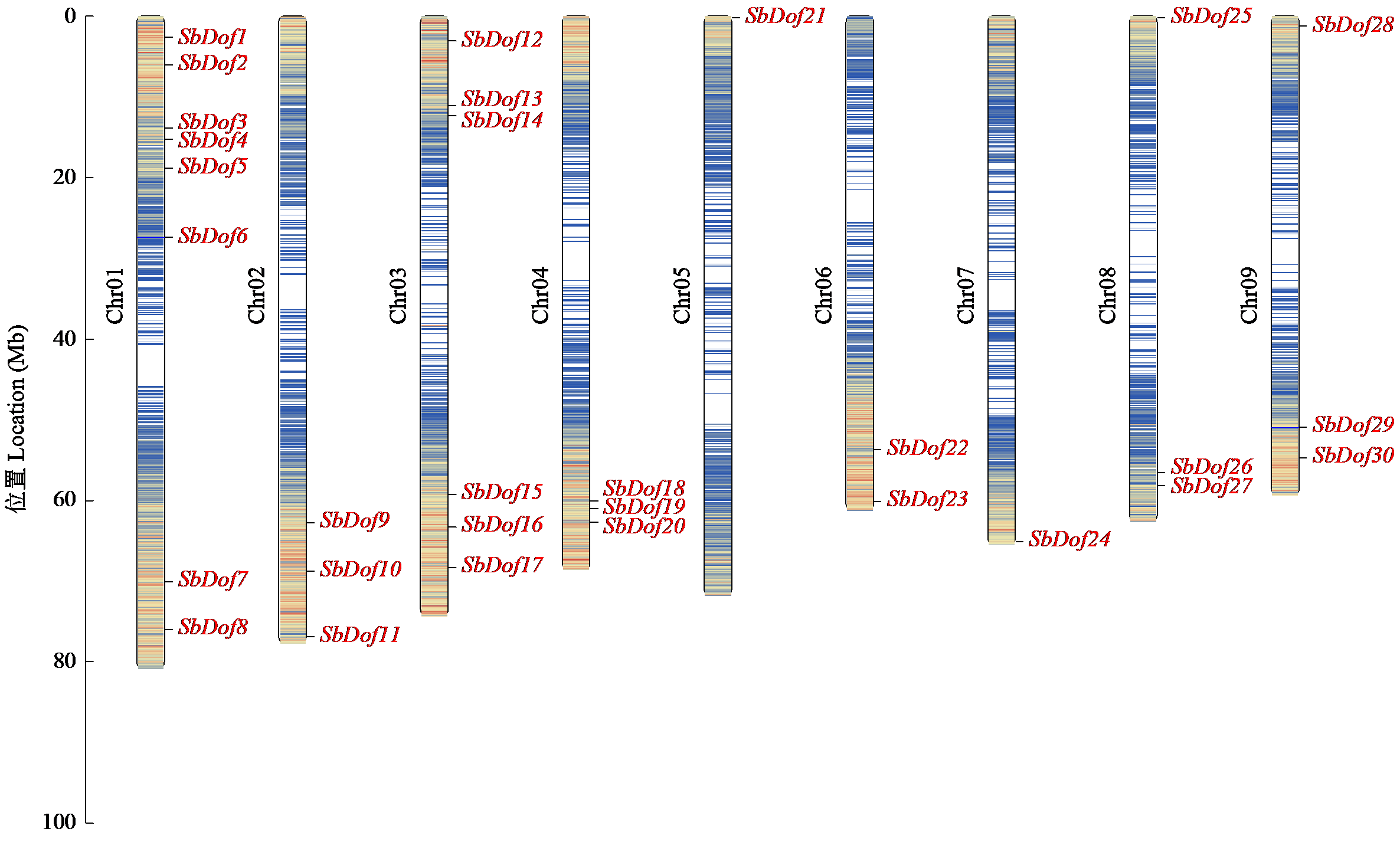
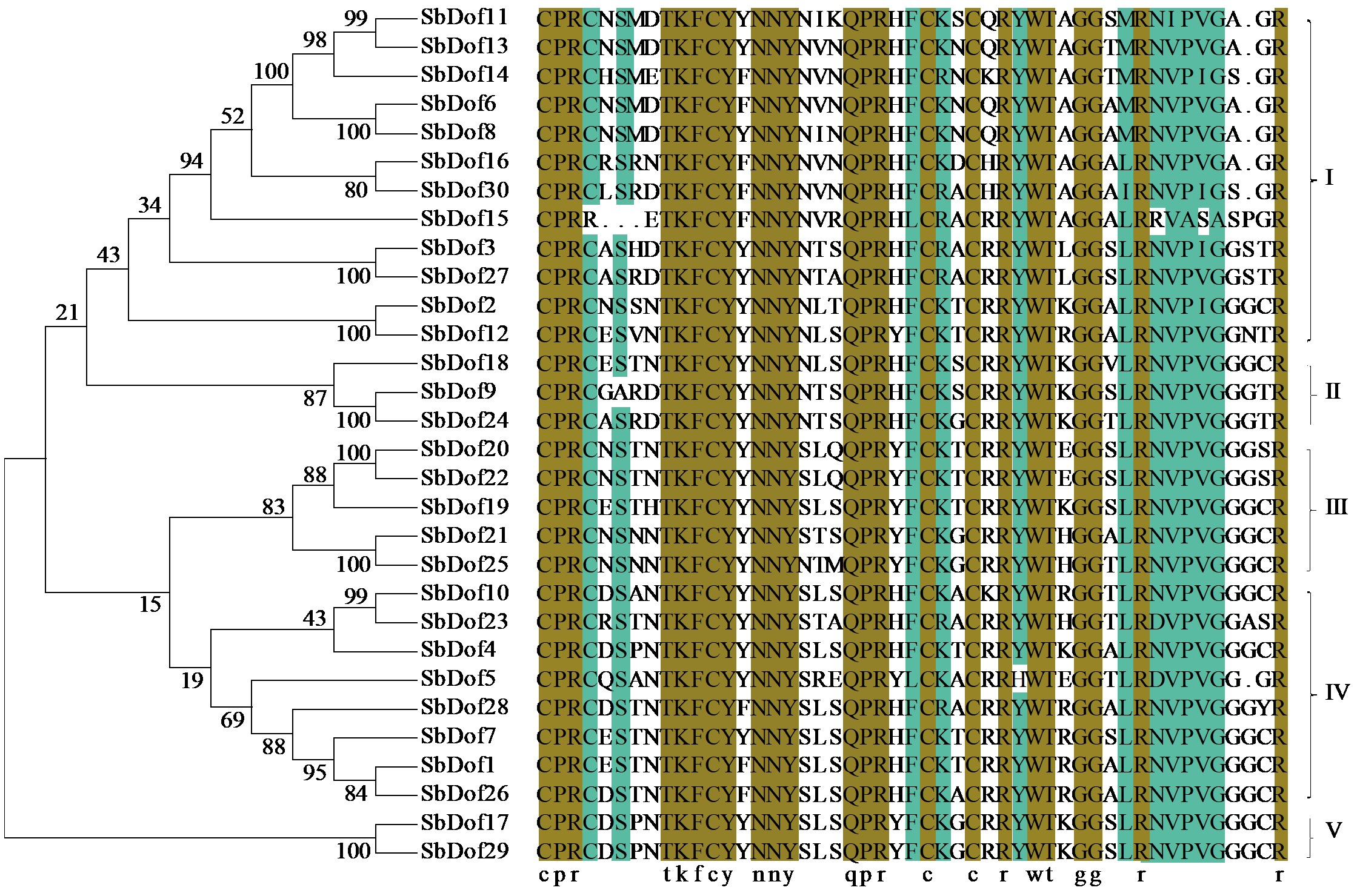
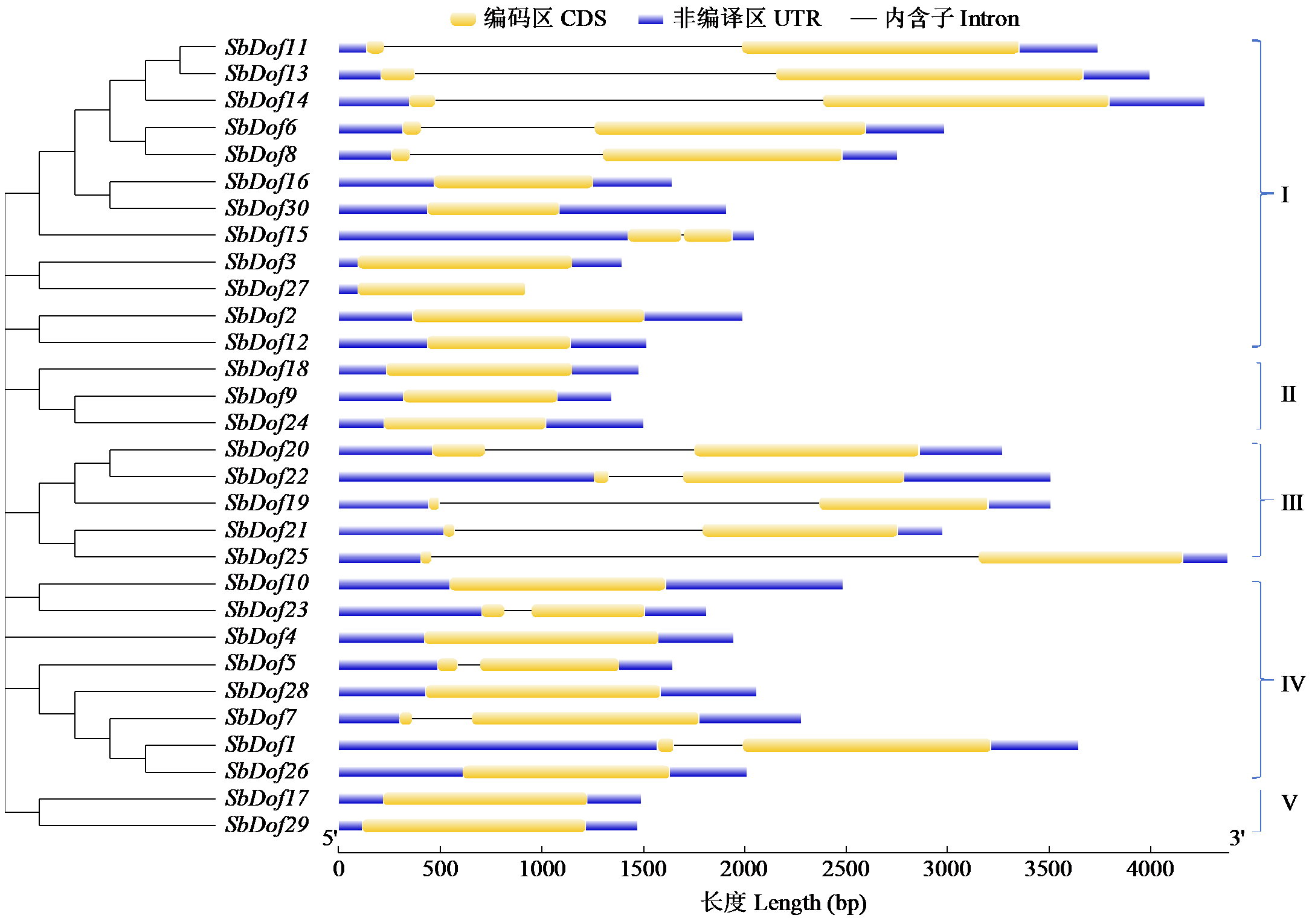
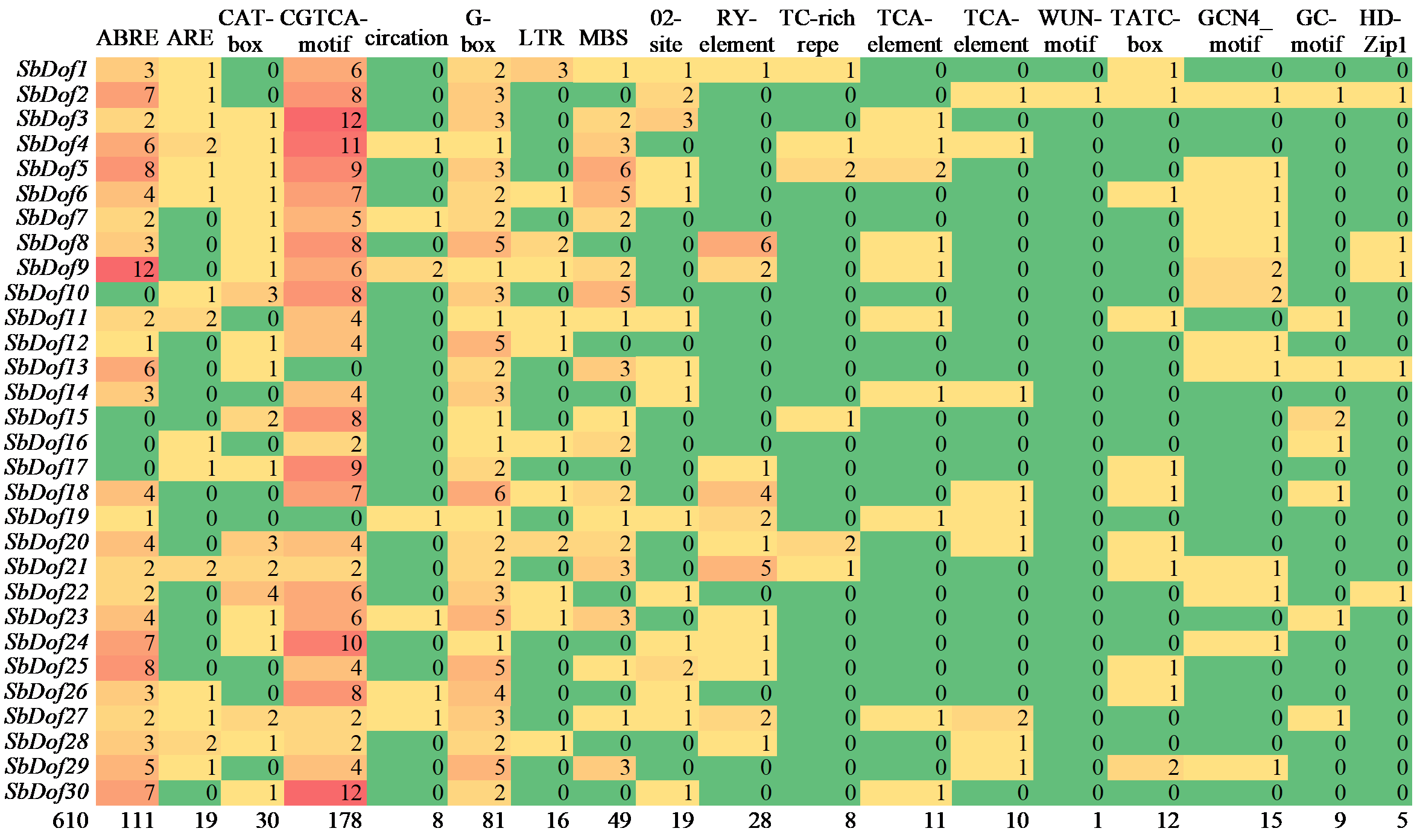
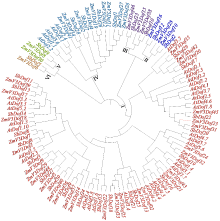
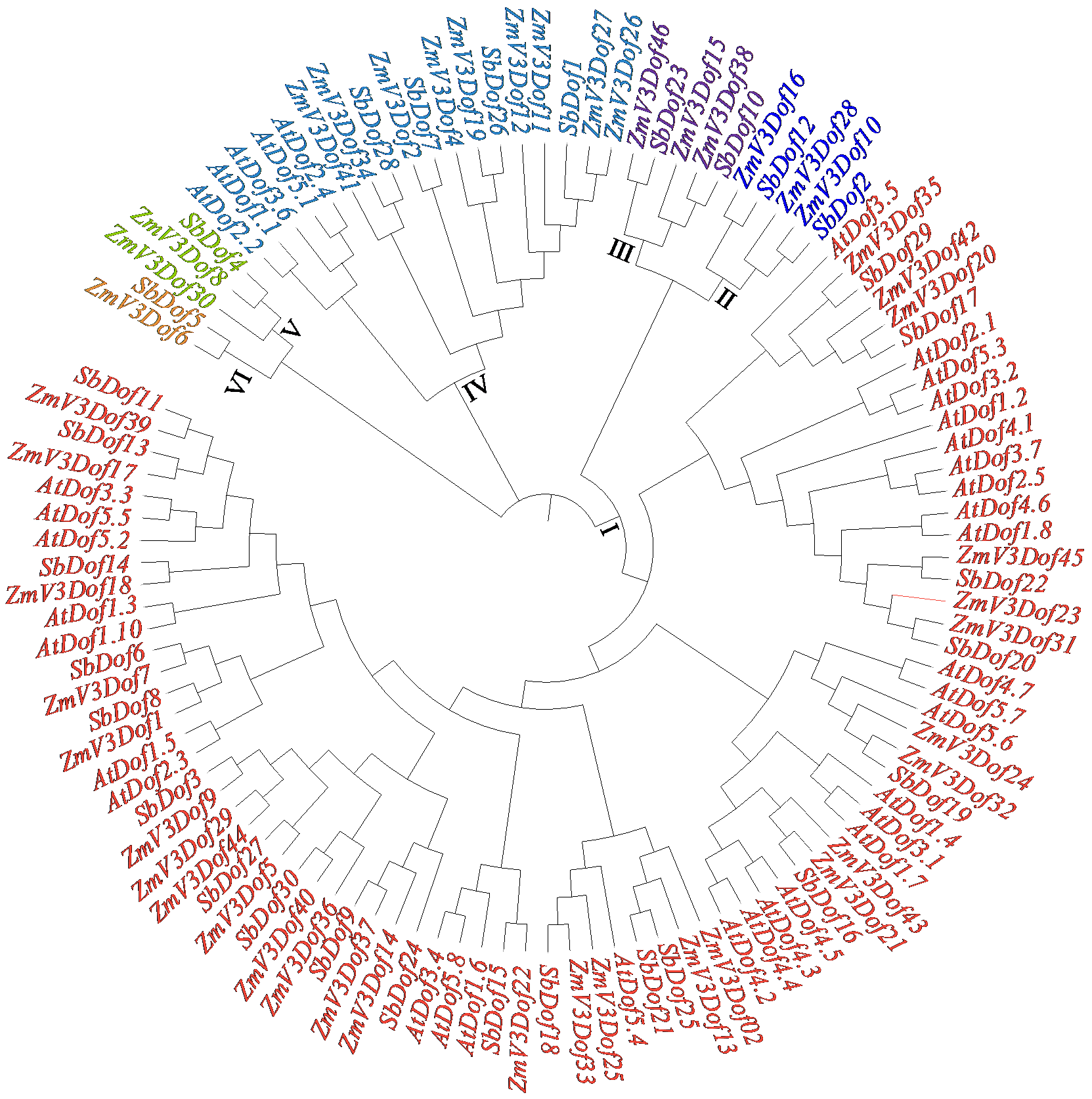
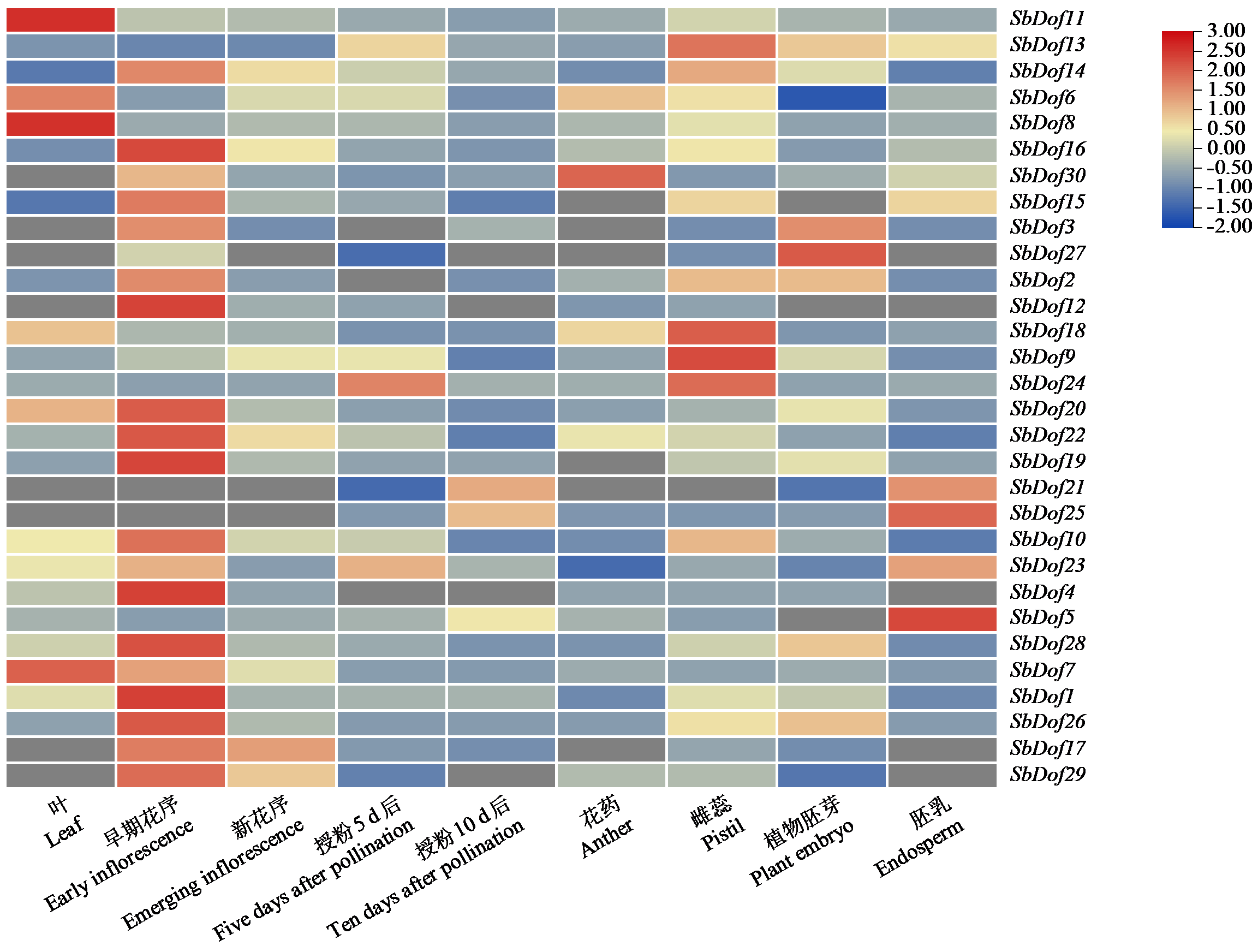
Identification and Expression Analysis of Dof Gene Family in Sorghum
Fu Senjie1( ), Song Yinghui1, Zhu Cancan1, Ye Zhenyan1, Qin Na1, Dai Shutao1, Liu Haixia2, Wang Chunyi1, Li Junxia1(
), Song Yinghui1, Zhu Cancan1, Ye Zhenyan1, Qin Na1, Dai Shutao1, Liu Haixia2, Wang Chunyi1, Li Junxia1( )
)
- 1Cereal Crops Institute, Henan Academy of Agricultural Sciences, Zhengzhou 450002, Henan, China
2Changge Agriculture and Rural Bureau, Xuchang 461500, Henan, China
| [1] | 邹剑秋, 王艳秋, 柯福来, 等. 高粱产业发展现状及前景展望. 山西农业大学学报(自然科学版), 2020, 40(3):2-8. |
| [2] | 姚茂星, 周光怡, 任明见, 等. 高粱GATA转录因子家族的鉴定和表达模式分析. 分子植物育种, 2022, 20(10):3178-3187. |
| [3] |
杜巧丽, 蒋君梅, 陈美晴, 等. 高粱转录因子SbWRKY71基因的克隆及其在逆境胁迫下的表达分析. 核农学报, 2021, 35(7):1532-1539.
doi: 10.11869/j.issn.100-8551.2021.07.1532 |
| [4] | Li Z Y, Chao J T, Li X X, et al. Systematic analysis of the bZIP family in tobacco and functional characterization of NtbZIP 62 involvement in salt stress. Agronomy, 2021, 11(1):1-17. |
| [5] | Yanagisawa S, Schmidt R J. Diversity and similarity among recognition sequences of Dof transcription factors. The Plant Journal, 1999, 17(2):209-214. |
| [6] | Cominelli E, Galbiati M, Albertini A, et al. DOF-binding sites additively contribute to guard cell-specificity of AtMYB60 promoter. BMC Plant Biology, 2011, 11(1):162. |
| [7] | Yang J, Yang M F, Zhang W P, et al. A putative flowering- time-related Dof transcription factor gene, JcDof3, is controlled by the circadian clock in Jatropha curcas. Plant Science, 2011, 181 (6):667-674. |
| [8] | 李娅, 丁文杰, 江海燕, 等. Dof基因家族调节植物生长发育功能的研究进展. 西北植物学报, 2018, 38(9):1758-1766. |
| [9] | 王新亮, 彭玲, 王健, 等. 苹果Dof转录因子生物信息学及其表达分析. 江苏农业学报, 2021, 37(2):480-492. |
| [10] | 孟文静, 张家琦, 赵康路, 等. 谷子Dof转录因子家族分析及功能预测. 分子植物育种, 2019, 174(4):1080-1089. |
| [11] |
Yanagisawa S. Dof domain proteins: plant-specific transcription factors associated with diverse phenomena unique to plants. Plant and Cell Physiology, 2004, 45(4):386-391.
doi: 10.1093/pcp/pch055 pmid: 15111712 |
| [12] | Zhang A D, Wang W Q, Yang T, et al. Transcriptome analysis identifies a zinc finger protein regulating starch degradation in kiwifruit. Plant Physiology, 2018, 178(2):850-863. |
| [13] | Shim Y, Kang K, An G, et al. Rice DNA-binding one zinc finger 24 (OsDOF24) delays leaf senescence in a jasmonate-mediated pathway. Plant & Cell Physiology, 2019, 60(9):2065-2076. |
| [14] |
Huang W, Huang Y, Li M Y, et al. Dof transcription factors in carrot: genome-wide analysis and their response to abiotic stress. Biotechnology Letters, 2016, 38(1):145-155.
doi: 10.1007/s10529-015-1966-2 pmid: 26466595 |
| [15] | 赵梦琪, 齐豫川, 雷振生, 等. 粗山羊草Dof转录因子家族基因的鉴定与分析. 分子植物育种, 2017, 15(7):2590-2597. |
| [16] | Su Y, Liang W, Liu Z J, et al. Overexpression of GhDof1 improved salt and cold tolerance and seed oil content in Gossypium hirsutum. Journal of Plant Physiology, 2017, 218:222-234. |
| [17] | Wu Q, Li D Y, Li D J, et al. Overexpression of OsDof12 affects plant architecture in rice (Oryza sativa L.). Frontiers in Plant Science, 2015, 6:833. |
| [18] | Chen W Q, Chao G, Singh K B. The promoter of a H2O2- inducible, Arabidopsis glutathione S-transferase gene contains closely linked OBF-and OBP1-binding sites. The Plant Journal, 1996, 10(6):955-966. |
| [19] | 张丹丹, 杨瑶君, 江纳, 等. 黄藤Dof家族的全基因组鉴定及系统进化分析. 西南林业大学学报(自然科学), 2021, 41(6):126-138. |
| [20] | Zhou Y, Cheng Y, Wan C P, et al. Genome-wide characterization and expression analysis of the Dof gene family related to abiotic stress in watermelon. PeerJ, 2020, 8(1/2):e8358. |
| [21] | 闵远昌, 叶钰澜, 胡欢, 等. 玉米Dof转录因子的全基因组鉴定和表达模式分析. 分子植物育种, 2024, 22(8):2347-2447. |
| [22] | 贾利强, 赵秋芳, 陈曙, 等. 9个玉米Dof基因对逆境胁迫的响应模式分析. 西北植物学报, 2020, 40(8):1347-1355. |
| [23] | 王泽民, 晋昕, 张飞燕, 等. Dof转录因子在作物逆境胁迫响应及农艺性状改良中的作用. 生物学杂志, 2023, 40(4):1736-2095. |
| [24] | 张凤娇. 拟南芥AtDof2.3基因抗逆功能研究. 哈尔滨:东北林业大学, 2015. |
| [25] | Liu L, Su C, Wang Y C, et al. ATDof5.8 protein is the up-stream regulator of ANAC069 and is responsive to abiotic stress. Biochimie, 2015, 110:17-24. |
| [26] | 贾利强, 赵秋芳, 陈曙, 等. 11个玉米DOF基因对逆境胁迫的表达模式分析. 西北农业学报, 2021, 30(7):1018-1027. |
| [27] |
张焕欣, 李国权, 杨惠栋, 等. 甜瓜Dof家族全基因组鉴定与表达分析. 园艺学报, 2019, 46(11):2176-2187.
doi: 10.16420/j.issn.0513-353x.2019-0115 |
| [28] |
葛敏, 吕远大, 李坦, 等. 玉米Dof转录因子家族的全基因组鉴定与分析. 中国农业科学, 2014, 47(23):4563-4573.
doi: 10.3864/j.issn.0578-1752.2014.23.002 |
| [29] | 赵振祥, 周东波, 张银霞, 等. 普通烟草Dof转录因子家族的全基因组鉴定及分析. 中国烟草科学, 2022, 43(4):70-77. |
| [30] | Corrale A R, Carrillo L, Lasierra P, et al. Multifaceted role of cycling DOF factor 3 (CDF3) in the regulation of flowering time and abiotic stress responses in Arabidopsis. Plant Cell and Environment, 2017, 40(5):748-764. |
| [31] | 唐跃辉, 包欣欣, 王健, 等. 小桐子Dof基因家族生物信息学与表达分析. 江苏农业学报, 2019, 35(1):15-25. |
| [32] | Yang Z, Chi X Y, Guo F F, et al. SbWRKY30 enhances the drought tolerance of plants and regulates a drought stress- responsive gene, SbRD19, in sorghum. Journal of Plant Physiology, 2020, 246:153142. |
| [33] | Tang Q, Zhang X D, Guo J, et al. Tomato SlPti5 plays a regulative role in the plant immune response against Botrytis cinerea through modulation of ROS system and hormone pathways. Journal of Integrative Agriculture, 2022, 21(3):697- 709. |
| [1] | Jiang Fenglin, Lei Xiongbiao, Zhao Jiaxuan, Huang Min, Li Manfei, Du Hewei. Growth and Development Process and Molecular Regulation Mechanism of Plant Root Hair [J]. Crops, 2024, 40(6): 9-17. |
| [2] | Yao Qi, Wang Hao, Xu Lingqing, Xing Wang, Liu Dali, Lu Zhenqiang. Genome-Wide Identification and Expression Analysis of C2H2 Zinc-Finger Protein Transcription Factor Family in Sugar Beet under Cadmium Stress [J]. Crops, 2024, 40(4): 33-42. |
| [3] | Lü Baolian, Yang Yuxin, Cui Licao, Shi Feng, Ma Liang, Kong Xiuying, Zhang Lichao, Ni Zhiyong. Identification of bHLH Family Transcription Factors of Wheat and Expression Analysis under Salt Stress [J]. Crops, 2024, 40(1): 65-72. |
| [4] | Jiang Yusong, Li Honglei, Li Zhexin, Xu Xiaoyu, Li Longyun, Huang Mengjun. Identification and Expression Analysis of the ZoWRKY Family in Stress Responses Based on Transcriptome Data of Ginger (Zingiber officinale Roscoe) [J]. Crops, 2022, 38(4): 37-45. |
| [5] | Wang Tong, Zhao Xiaodong, Zhen Pingping, Chen Jing, Chen Mingna, Chen Na, Pan Lijuan, Wang Mian, Xu Jing, Yu Shanlin, Chi Xiaoyuan, Zhang Jiancheng. Genome-Wide Identification and Characteristic Analyzation of the TCP Transcription Factors Family in Peanut [J]. Crops, 2021, 37(2): 35-44. |
| [6] | Li Guolong, Wu Haixia, Sun Yaqing. Construction of RNAi Expression Vector of BvWRKY23 Gene in Sugar Beet [J]. Crops, 2020, 36(5): 41-47. |
| [7] | Xu Yuanyuan, Zhao Peng, Hong Quanchun, Zhu Xiaoqin, Pei Dongli. Isolation and Expression Analysis of Transcription Factor Gene TaMYB70 in Wheat [J]. Crops, 2020, 36(4): 84-90. |
| [8] | Duan Junzhi, Qi Xueli, Feng Lili, Zhang Huifang, Sun Yan, Yan Zhaoling, Chen Haiyan, Qi Hongzhi, Fan Wenjie, Yang Cuiping, Liu Yuxia, Ren Yinling, Zhang Jiayuan, Li Ying, Zhuo Wenfei. Progress on Application of Drought Tolerance Genes in Wheat Drought Tolerance Genetic Engineering [J]. Crops, 2020, 36(3): 7-15. |
| [9] | Yang Junkai,Shen Yang,Cai Xiaoxi,Wu Shengyang,Li Jianwei,Sun Mingzhe,Jia Bowei,Sun Xiaoli. Genome-Wide Identification and Expression Patterns Analysis of the PHD Family Protein in Glycine max [J]. Crops, 2019, 35(3): 55-65. |
| [10] | Wu Hao,Li Yanmin,Xie Chuanxiao. Research Advances on Physiological Basis and Gene Discovery for Thermal Tolerance in Crops [J]. Crops, 2018, 34(5): 1-9. |
| [11] | Xue Zhang,Yuejia Yin,Bei Fan,Huijie Li,Xiaoyu Fei,Xiyan Cui. Advances on the Structural Characteristics and Function of Dof Transcription Factors in Plant [J]. Crops, 2016, 32(2): 14-20. |
|