作物杂志,2024, 第1期: 31–39 doi: 10.16035/j.issn.1001-7283.2024.01.005
所属专题: 玉米专题
玉米穗粗一般配合力多位点全基因组关联分析和基因组预测
- 河南省农业科学院粮食作物研究所,450002,河南郑州
Multi-Locus Genome-Wide Association Study and Genomic Prediction for General Combining Ability of Maize Ear Diameter
Ma Juan( ), Huang Lu, Yu Ting, Guo Guojun, Zhu Weihong, Liu Jingbao
), Huang Lu, Yu Ting, Guo Guojun, Zhu Weihong, Liu Jingbao
- Institute of Cereal Crops, Henan Academy of Agricultural Sciences, Zhengzhou 450002, Henan, China
摘要:
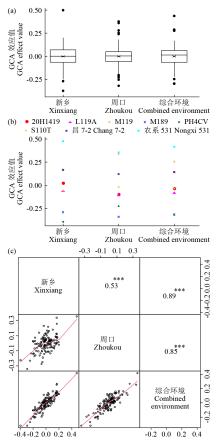
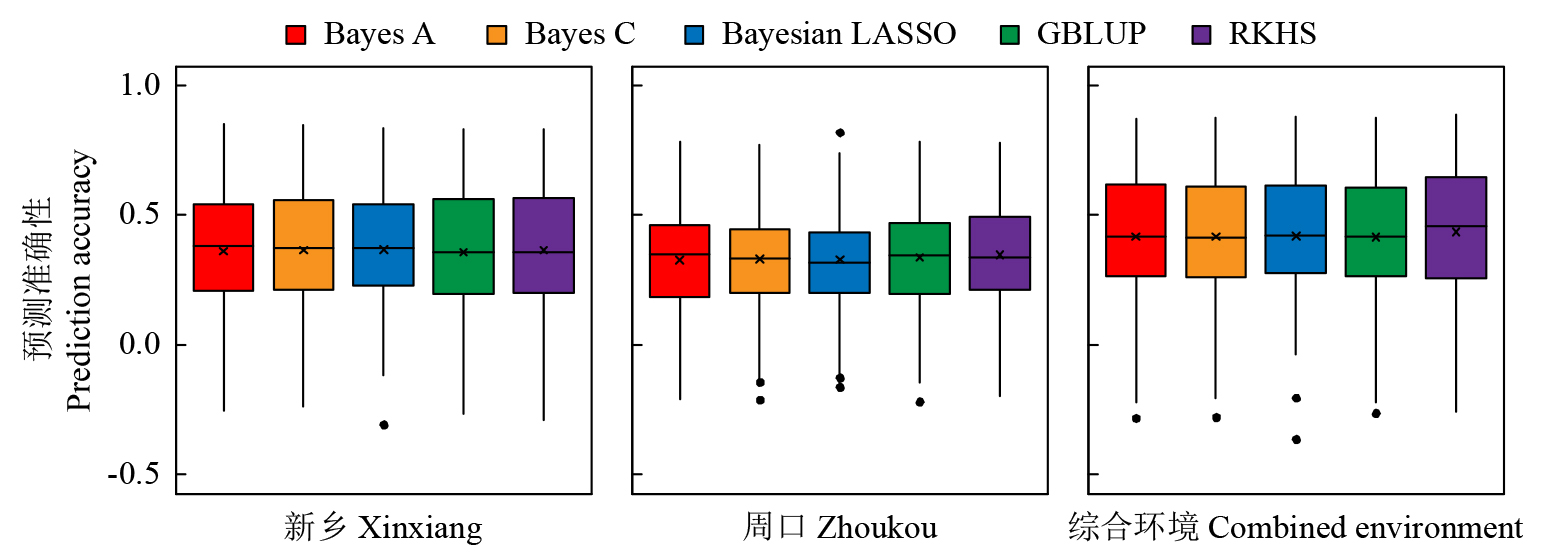
穗粗是一个重要的穗部性状,一般配合力(GCA)是杂交育种中衡量亲本组配能力的一个重要指标。以NCII遗传交配设计组配的537个F1杂交组合为材料,利用7种多位点全基因组关联分析方法(MGWAS)对河南新乡、周口和综合环境穗粗GCA开展定位分析,并研究MGWAS方法挖掘的显著关联位点作为固定效应对穗粗GCA预测准确性的影响,为穗粗GCA关键位点的基因组选择(GS)育种利用提供指导。结果表明,共检测到18个穗粗GCA显著关联SNP(P<8.52E-07),其中6个位点利用2~5种MGWAS方法同时检测到,3个位点在2个环境中均检测到。5种GS随机效应模型对3个环境穗粗GCA预测准确性均较低,为0.32~0.44。3个环境中,显著位点作为固定效应加入预测模型中均能有效地提高穗粗GCA基因组预测能力,可将预测精度提高到0.38~0.64。
| [1] |
Su C F, Wang W, Gong S L, et al. High density linkage map construction and mapping of yield trait QTLs in maize (Zea mays) using the genotyping-by-sequencing (GBS) technology. Frontiers in Plant Science, 2017, 8:706-719.
doi: 10.3389/fpls.2017.00706 |
| [2] | 监立强. 玉米产量相关性状及其一般配合力的关联分析. 保定:河北农业大学, 2017. |
| [3] | 刘文童, 监立强, 郭晋杰, 等. 玉米穗部性状及其一般配合力的关联分析. 植物遗传资源学报, 2020, 21(3):706-715. |
| [4] |
Wang J B, Zhang Z W. GAPIT Version 3: Boosting power and accuracy for genomic association and prediction. Genomics Proteomics Bioinformatics, 2021, 19(4):629-640.
doi: 10.1016/j.gpb.2021.08.005 |
| [5] |
Zhang Y W, Tamba C L, Wen Y J, et al. mrMLM v4.0.2: An R platform for multi-locus genome-wide association studies. Genomics Proteomics Bioinformatics, 2020, 18(4):481-487.
doi: 10.1016/j.gpb.2020.06.006 |
| [6] |
He J B, Meng S, Zhao T J, et al. An innovative procedure of genome-wide association analysis fits studies on germplasm population and plant breeding. Theoretical and Applied Genetics, 2017, 130(11):2327-2343.
doi: 10.1007/s00122-017-2962-9 pmid: 28828506 |
| [7] |
Ma J, Cao Y Y. Genetic dissection of grain yield of maize and yield-related traits through association mapping and genomic prediction. Frontiers in Plant Science, 2021, 12:690059-690069.
doi: 10.3389/fpls.2021.690059 |
| [8] |
Ma J, Wang L F, Cao Y Y, et al. Association mapping and transcriptome analysis reveal the genetic architecture of maize kernel size. Frontiers in Plant Science, 2021, 12:632788-632799.
doi: 10.3389/fpls.2021.632788 |
| [9] |
Malik P, Kumar J, Sharma S, et al. Multi-locus genome-wide association mapping for spike-related traits in bread wheat (Triticum aestivum L.). BMC Genomics, 2021, 22(1):597-617.
doi: 10.1186/s12864-021-07834-5 |
| [10] |
Su J J, Wang C X, Hao F S, et al. Genetic detection of lint percentage applying single-locus and multi-locus genome-wide association studies in Chinese early-maturity upland cotton. Frontiers in Plant Science, 2019, 10:964-974.
doi: 10.3389/fpls.2019.00964 pmid: 31428110 |
| [11] |
Meuwissen T H, Hayes B J, Goddard M E. Prediction of total genetic value using genome-wide dense marker maps. Genetics, 2001, 157(4):1819-1829.
doi: 10.1093/genetics/157.4.1819 pmid: 11290733 |
| [12] |
Park T, Casella G. The Bayesian Lasso. Journal of the American Statistical Association, 2008, 103(482):681-686.
doi: 10.1198/016214508000000337 |
| [13] |
Gianola D, van Kaam J B C H M. Reproducing kernel Hilbert spaces regression methods for genomic assisted prediction of quantitative traits. Genetics, 2008, 178(4):2289-2303.
doi: 10.1534/genetics.107.084285 pmid: 18430950 |
| [14] |
Guo Z, Tucker D M, Lu J, et al. Evaluation of genome-wide selection efficiency in maize nested association mapping populations. Theoretical and Applied Genetics, 2012, 124(2):261-275.
doi: 10.1007/s00122-011-1702-9 pmid: 21938474 |
| [15] |
Lian L, Jacobson A, Zhong S. Genome wide prediction accuracy within 969 maize biparental populations. Crop Science, 2014, 54(4):1514-1522.
doi: 10.2135/cropsci2013.12.0856 |
| [16] |
Liu X G, Wang H W, Wang H, et al. Factors affecting genomic selection revealed by empirical evidence in maize. The Crop Journal, 2018, 6(4):341-352.
doi: 10.1016/j.cj.2018.03.005 |
| [17] |
Liu X G, Wang H W, Hu X J, et al. Improving genomic selection with quantitative trait loci and nonadditive effects revealed by empirical evidence in maize. Frontiers in Plant Science, 2019, 10:1129-1141.
doi: 10.3389/fpls.2019.01129 pmid: 31620155 |
| [18] |
Sitonik C, Suresh L M, Beyene Y, et al. Genetic architecture of maize chlorotic mottle virus and maize lethal necrosis through GWAS, linkage analysis and genomic prediction in tropical maize germplasm. Theoretical and Applied Genetics, 2019, 132 (8):2381-2399.
doi: 10.1007/s00122-019-03360-x pmid: 31098757 |
| [19] |
Yuan Y, Cairns J E, Babu R, et al. Genome-wide association mapping and genomic prediction analyses reveal the genetic architecture of grain yield and flowering time under drought and heat stress conditions in maize. Frontiers in Plant Science, 2019, 9:1919-1933.
doi: 10.3389/fpls.2018.01919 |
| [20] |
Bian Y, Holland J B. Enhancing genomic prediction with genome-wide association studies in multiparental maize populations. Heredity, 2017, 118(6):585-593.
doi: 10.1038/hdy.2017.4 pmid: 28198815 |
| [21] |
Spindel J E, Begum H, Akdemir D, et al. Genome-wide prediction models that incorporate de novo GWAS are a powerful new tool for tropical rice improvement. Heredity, 2016, 116(4):395-408.
doi: 10.1038/hdy.2015.113 pmid: 26860200 |
| [22] |
Arruda M, Lipka A, Brown P, et al. Comparing genomic selection and marker-assisted selection for Fusarium head blight resistance in wheat (Triticum aestivum). Molecular Breeding, 2016, 36(7):1-11.
doi: 10.1007/s11032-015-0425-z |
| [23] |
Herter C P, Ebmeyer E, Kollers S, et al. Accuracy of within- and among-family genomic prediction for Fusarium head blight and Septoria tritici blotch in winter wheat. Theoretical and Applied Genetics, 2019, 132(4):1121-1135.
doi: 10.1007/s00122-018-3264-6 |
| [24] |
Michel S, Kummer C, Gallee M, et al. Improving the baking quality of bread wheat by genomic selection in early generations. Theoretical and Applied Genetics, 2018, 131(2):477-493.
doi: 10.1007/s00122-017-2998-x pmid: 29063161 |
| [25] |
Moore J K, Manmathan H K, Anderson V A, et al. Improving genomic prediction for pre-harvest sprouting tolerance in wheat by weighting large-effect quantitative trait loci. Crop Science, 2017, 57(3):1315-1324.
doi: 10.2135/cropsci2016.06.0453 |
| [26] |
Zhao Y, Mette M F, Gowda M, et al. Bridging the gap between marker-assisted and genomic selection of heading time and plant height in hybrid wheat. Heredity, 2014, 112(6):638-645.
doi: 10.1038/hdy.2014.1 pmid: 24518889 |
| [27] |
Pérez P, de los Campos G,. Genome-wide regression and prediction with the BGLR statistical package. Genetics, 2014, 198(2):483-495.
doi: 10.1534/genetics.114.164442 pmid: 25009151 |
| [28] |
Wen Y J, Zhang H, Ni Y L, et al. Methodological implementation of mixed linear models in multi-locus genome-wide association studies. Briefings in Bioinformatics, 2018, 19(4):700-712.
doi: 10.1093/bib/bbw145 |
| [29] |
Zhang J, Feng J Y, Ni Y L, et al. pLARmEB:integration of least angle regression with empirical Bayes for multilocus genome- wide association studies. Heredity, 2017, 118(6):517-524.
doi: 10.1038/hdy.2017.8 pmid: 28295030 |
| [30] |
Zhou G F, Zhu Q L, Mao Y X, et al. Multi-locus genome-wide association study and genomic selection of kernel moisture content at the harvest stage in maize. Frontiers in Plant Science, 2021, 12:697688-697700.
doi: 10.3389/fpls.2021.697688 |
| [31] |
Cui Y R, Zhang F, Zhou Y L. The application of multi-locus GWAS for the detection of salt-tolerance loci in rice. Frontiers in Plant Science, 2018, 9:1464-1472.
doi: 10.3389/fpls.2018.01464 pmid: 30337936 |
| [32] |
Zhou Z P, Li G L, Tan S Y, et al. A QTL atlas for grain yield and its component traits in maize (Zea mays). Plant Breeding, 2020, 139(3):562-574.
doi: 10.1111/pbr.v139.3 |
| [33] |
Lu X, Zhou Z Q, Yuan Z H, et al. Genetic dissection of the general combining ability of yield-related traits in maize. Frontiers in Plant Science, 2020, 11:788-802.
doi: 10.3389/fpls.2020.00788 pmid: 32793248 |
| [34] |
Zhang X X, Guan Z R, Li Z L, et al. A combination of linkage mapping and GWAS brings new elements on the genetic basis of yield-related traits in maize across multiple environments. Theoretical and Applied Genetics, 2020, 133(10):2881-2895.
doi: 10.1007/s00122-020-03639-4 pmid: 32594266 |
| [35] |
Chen L, An Y, Li Y X, et al. Candidate loci for yield-related traits in maize revealed by a combination of MetaQTL analysis and regional association mapping. Frontiers in Plant Science, 2017, 8:2190-2203.
doi: 10.3389/fpls.2017.02190 pmid: 29312420 |
| [36] |
Brooks L, Strable J, Zhang X, et al. Microdissection of shoot meristem functional domains. PLoS Genetics, 2009, 5(5):e1000476.
doi: 10.1371/journal.pgen.1000476 |
| [37] |
Zou C S, Jiang W B, Yu D Q. Male gametophyte-specific WRKY34 transcription factor mediates cold sensitivity of mature pollen in Arabidopsis. Journal of Experimental Botany, 2010, 61(14):3901-3914.
doi: 10.1093/jxb/erq204 pmid: 20643804 |
| [38] |
Wang Y, Li Y, He S P, et al. A cotton (Gossypium hirsutum) WRKY transcription factor (GhWRKY22) participates in regulating anther/pollen development. Plant Physiology and Biochemistry, 2019, 141:231-239.
doi: S0981-9428(19)30240-2 pmid: 31195253 |
| [39] |
Yao J, Zhao D H, Chen X M, et al. Use of genomic selection and breeding simulation in cross prediction for improvement of yield and quality in wheat (Triticum aestivum L.). The Crop Journal, 2018, 6(4):353-365.
doi: 10.1016/j.cj.2018.05.003 |
| [40] |
Ali M, Zhang Y, Rasheed A, et al. Genomic prediction for grain yield and yield-related traits in Chinese winter wheat. International Journal of Molecular Sciences, 2020, 21(4):1342-1359.
doi: 10.3390/ijms21041342 |
| [41] |
马娟, 曹言勇, 朱卫红. 玉米穗轴粗和出籽率全基因组预测分析. 植物遗传资源学报, 2021, 22(6):1708-1715.
doi: 10.13430/j.cnki.jpgr. 20210405002 |
| [42] |
Heffner E L, Lorenz A J, Jannink J L, et al. Plant breeding with genomic selection:gain per unit time and cost. Crop Science, 2010, 50(5):1681-1690.
doi: 10.2135/cropsci2009.11.0662 |
| [1] | 冯勇, 侯旭光, 薛春雷, 张来厚, 宋国栋, 苏敏莉, 傅晓华, 孙宇燕. 内蒙古玉米品种适宜生态区划分[J]. 作物杂志, 2024, (1): 23–30 |
| [2] | 王海涛, 任春梅, 董岩, 李硕, 程兆榜, 季英华. 江苏淮安市高粱上玉米黄花叶病毒的分子检测与鉴定[J]. 作物杂志, 2024, (1): 233–238 |
| [3] | 毋莹, 胡蝶, 李婷, 段乾元, 韦宁宁, 张兴华, 徐淑兔, 薛吉全. 玉米WRKY转录因子IIc亚家族分析及其在干旱胁迫下的表达分析[J]. 作物杂志, 2024, (1): 80–89 |
| [4] | 金玉, 郭新宇, 张颖, 李大壮, 王璟璐. 玉米叶片气孔表型鉴定及研究进展[J]. 作物杂志, 2023, (6): 1–10 |
| [5] | 吴琦, 明博, 高尚, 杨宏业, 张川, 初振东, 李少昆. 东北冷凉区玉米籽粒脱水模型构建策略研究[J]. 作物杂志, 2023, (6): 108–113 |
| [6] | 梁忠宇, 薛军, 张国强, 明博, 沈东萍, 方梁, 周林立, 张玉芹, 杨恒山, 王克如, 李少昆. 水肥一体化施磷量对玉米抗倒伏能力的影响[J]. 作物杂志, 2023, (6): 190–194 |
| [7] | 曹庆军, 李刚, 杨浩, 娄玉勇, 杨粉团, 孔凡丽, 李辛琲, 赵新凯, 姜晓莉. 不同耕作方式对春玉米种床质量的影响及其与幼苗群体建成和产量的关系[J]. 作物杂志, 2023, (5): 249–254 |
| [8] | 于乐, 李林, 黄红娟, 黄兆峰, 朱文达, 魏守辉. 湖北省玉米田杂草种类组成及群落特征[J]. 作物杂志, 2023, (5): 272–279 |
| [9] | 杨宗莹, 肖贵, 张红伟. 利用玉米F1群体进行玉米全株鲜重的全基因组预测分析[J]. 作物杂志, 2023, (5): 43–48 |
| [10] | 曲海涛, 李忠南, 王越人, 马艺文, 相洋, 邬生辉, 谭倬, 王纯, 魏强, 罗瑶, 李光发. 玉米百粒重遗传育种效应研究[J]. 作物杂志, 2023, (5): 66–70 |
| [11] | 杨密, 王美娟, 许海涛. 不同生态区玉米自交系苞叶动态发育差异性研究[J]. 作物杂志, 2023, (5): 81–90 |
| [12] | 袁刘正, 王会强, 王秋岭, 朱世蝶, 赵月强, 袁曼曼, 王会涛, 张运栋, 柳家友, 袁永强. 遮阴条件下玉米自交系配合力与遗传效应分析[J]. 作物杂志, 2023, (4): 104–109 |
| [13] | 郑飞, 陈静, 崔亚坤, 孔令杰, 孟庆长, 李杰, 刘瑞响, 张美景, 赵文明, 陈艳萍. 淮北不同生态区丰产稳产宜机收玉米新品种筛选[J]. 作物杂志, 2023, (4): 110–117 |
| [14] | 王丽萍, 白岚方, 王天昊, 王宵璇, 白云鹤, 王玉芬. 不同施氮水平对青贮玉米植株氮素积累和转运的影响[J]. 作物杂志, 2023, (4): 165–173 |
| [15] | 李雨新, 卢敏, 赵久然, 王荣焕, 徐田军, 吕天放, 蔡万涛, 张勇, 薛洪贺, 刘月娥. 京津唐夏玉米区生产现状调研分析[J]. 作物杂志, 2023, (4): 174–181 |
|
||