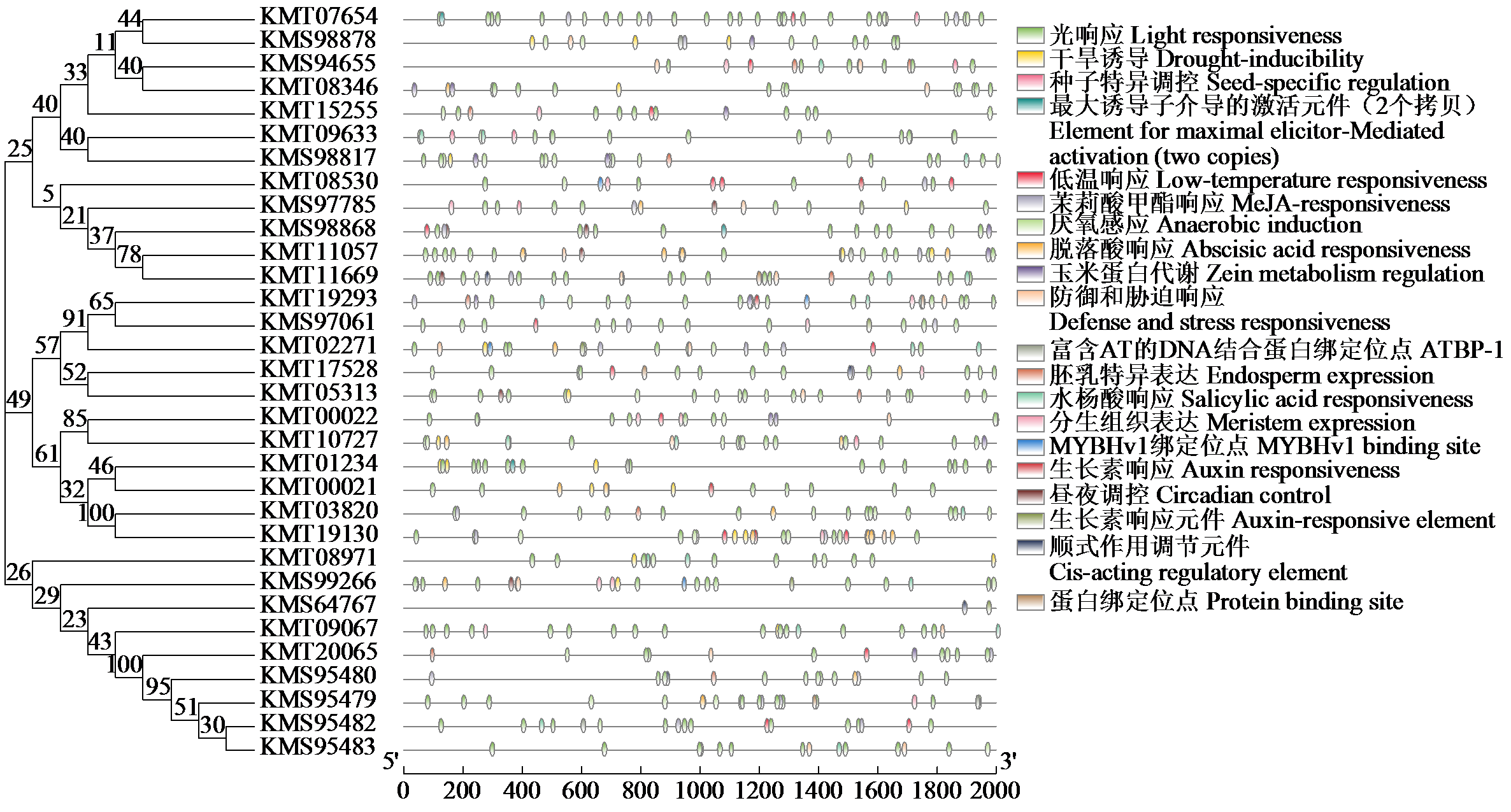
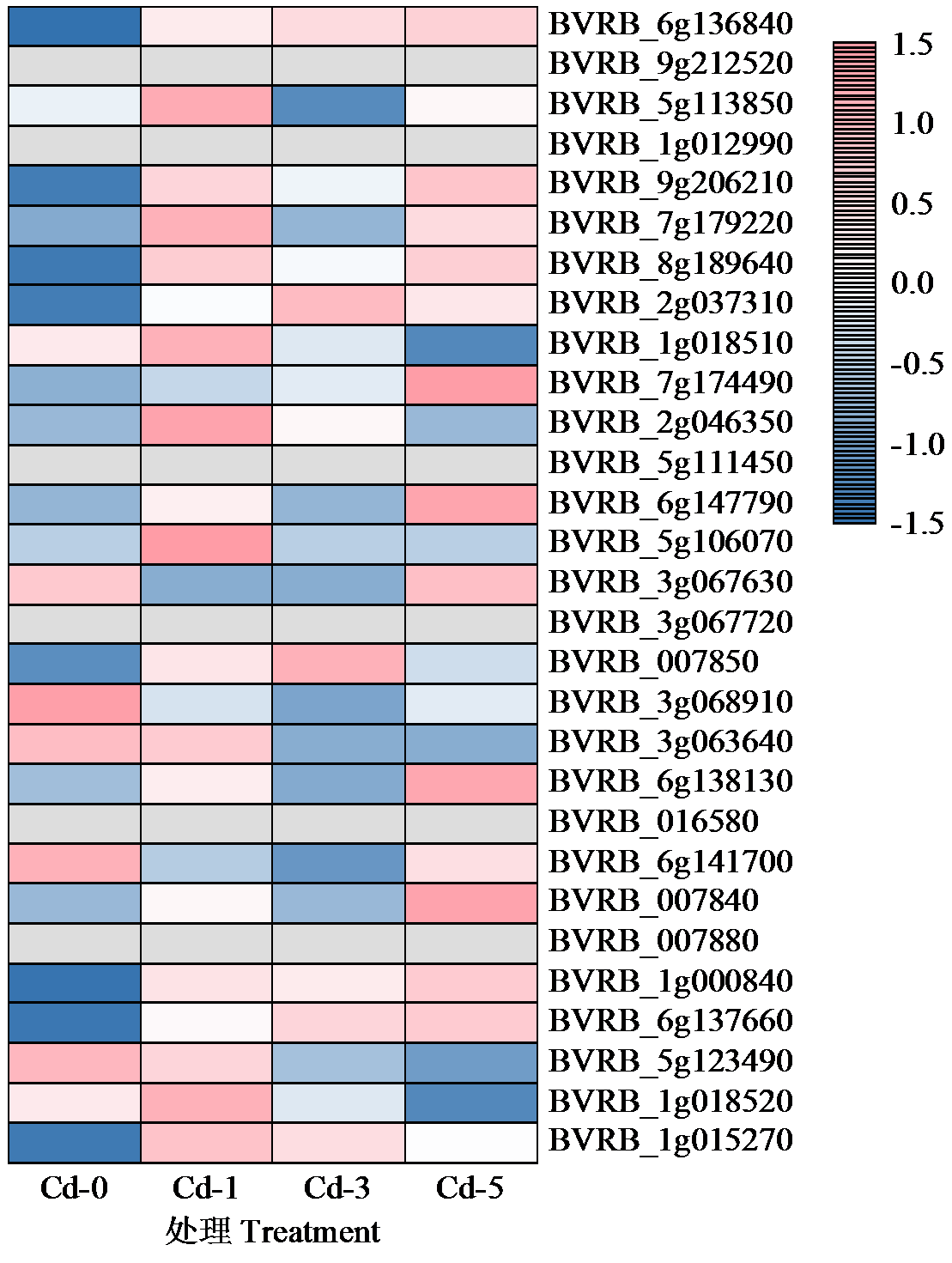
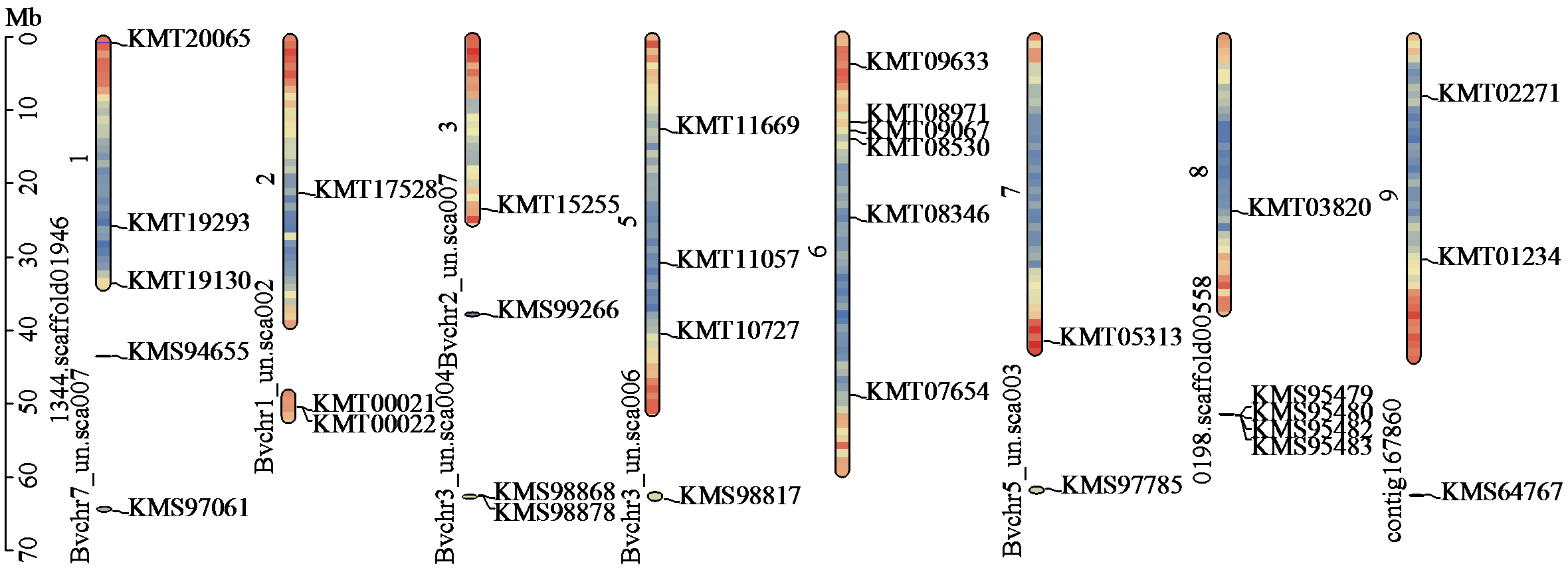Crops ›› 2024, Vol. 40 ›› Issue (4): 33-42.doi: 10.16035/j.issn.1001-7283.2024.04.005
Previous Articles Next Articles
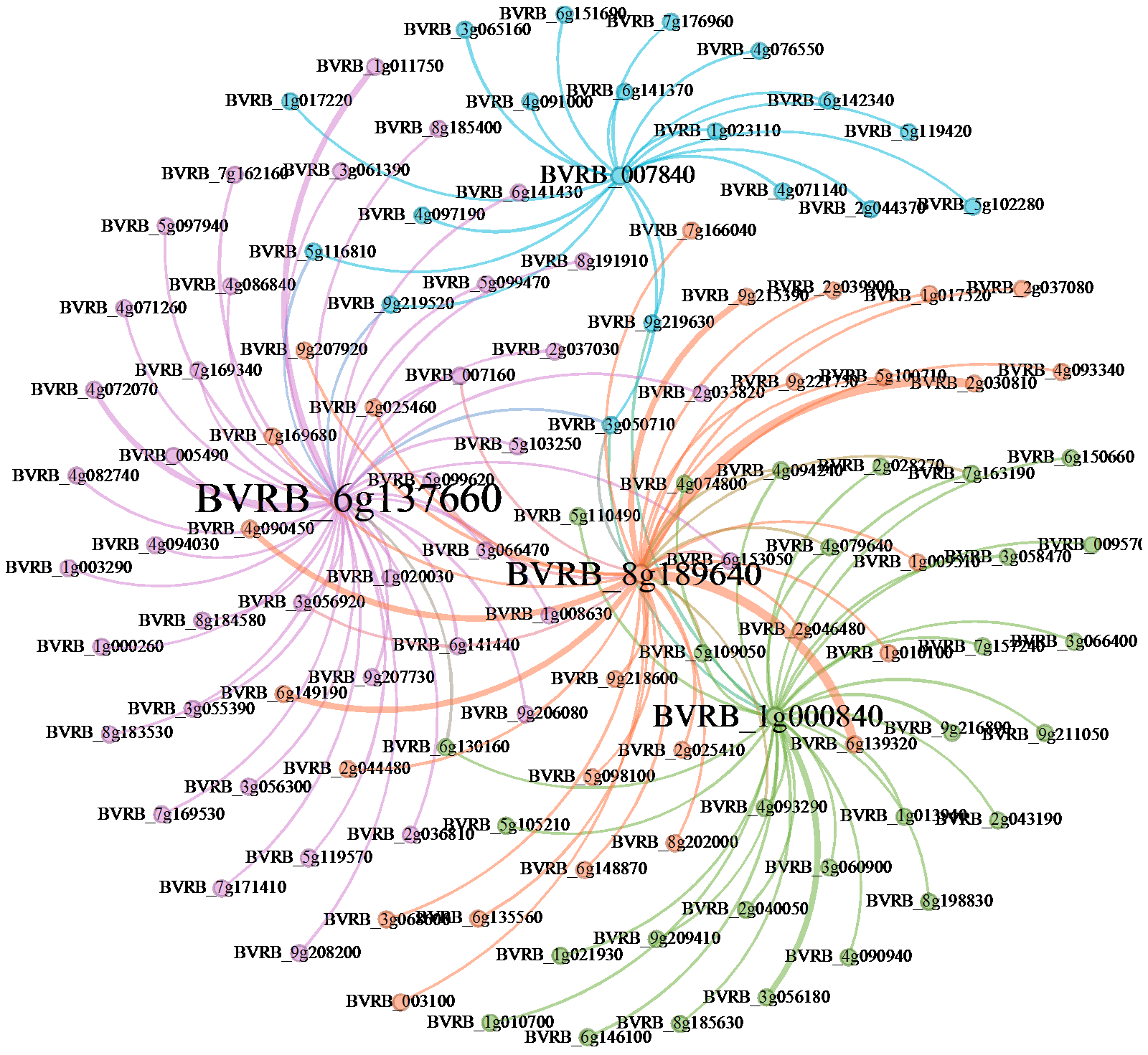
Genome-Wide Identification and Expression Analysis of C2H2 Zinc-Finger Protein Transcription Factor Family in Sugar Beet under Cadmium Stress
Yao Qi1,2,3( ), Wang Hao2,3, Xu Lingqing2,3, Xing Wang2,3, Liu Dali2,3, Lu Zhenqiang1(
), Wang Hao2,3, Xu Lingqing2,3, Xing Wang2,3, Liu Dali2,3, Lu Zhenqiang1( )
)
- 1Province Key Laboratory of Plant Gene and Biological Fermentation in Cold Regions / College of Life Sciences, Heilongjiang University, Harbin 150080, Heilongjiang, China
2National Beet Medium-Term Gene Bank / College of Advanced Agriculture and Ecological Environment, Heilongjiang University, Harbin 150080, Heilongjiang, China
3Key Laboratory of Sugar Beet Genetics and Breeding / College of Advanced Agriculture and Ecological Environment, Heilongjiang University, Harbin 150080, Heilongjiang, China
| [1] | Cui H Y, Chen J Q, Liu M J, et al. Genome-wide analysis of C2H2 zinc-finger gene family and its response to cold and drought stress in sorghum [Sorghum bicolor (L.) Moench]. International Journal of Molecular Sciences, 2022, 23(10):55-71. |
| [2] | Han G L, Lu C X, Guo J R, et al. C2H2 zinc-finger proteins:Master regulators of abiotic stress responses in plants. Frontiers in Plant Science, 2020, 11:115. |
| [3] |
Englbrecht C C, Schoof H, Böhm S. Conservation, diversification and expansion of C2H2 zinc-finger proteins in the Arabidopsis thaliana genome. BMC Genomics, 2004, 5(1):39.
doi: 10.1186/1471-2164-5-39 pmid: 15236668 |
| [4] |
Agarwal P, Arora R, Ray S, et al. Genome-wide identification of C2H 2 zinc-finger gene family in rice and their phylogeny and expression analysis. Plant Molecular Biology, 2007, 65(4):467-485.
doi: 10.1007/s11103-007-9199-y pmid: 17610133 |
| [5] |
Weng L, Zhao F F, Li R, et al. The zinc-finger transcription factor SlZFP2 negatively regulates abscisic acid biosynthesis and fruit ripening in tomato. Plant Physiology, 2015, 167(3):931-949.
doi: 10.1104/pp.114.255174 pmid: 25637453 |
| [6] | Liu Q G, Wang Z C, Xu X M, et al. Genome-wide analysis of C2H 2 zinc-finger family transcription factors and their responses to abiotic stresses in poplar (Populus trichocarpa). PLoS ONE, 2015, 10(8):e134753. |
| [7] |
Li Y, Chu Z N, Luo J Y, et al. The C2H2 zinc-finger protein SlZF3 regulates AsA synthesis and salt tolerance by interacting with CSN5B. Plant Biotechnology Journal, 2018, 16(6):1201-1213.
doi: 10.1111/pbi.12863 pmid: 29193661 |
| [8] | Lyu T Q, Liu W M, Hu Z W, et al. Molecular characterization and expression analysis reveal the roles of Cys2/His2 zinc-finger transcription factors during flower development of Brassica rapa subsp. chinensis. Plant Molecular Biology, 2020, 102(1/2):123-141. |
| [9] | 井建玲, 张鹏, 王振宇, 等. 木薯C2H2型锌指蛋白转录因子家族全基因组鉴定及表达分析. 植物生理学报, 2020, 56(12):2664-2676. |
| [10] | Liu Z, Coulter J A, Li Y, et al. Genome-wide identification and analysis of the Q-type C2H2 gene family in potato (Solanum tuberosum L.). International Journal of Biological Macromolecules, 2020, 153:327-340. |
| [11] | 李琳, 丁峰, 潘介春, 等. 植物锌指蛋白转录因子家族研究进展. 热带农业科学, 2020, 40(2):65-75. |
| [12] | Zhang C, Tong C, Cao L, et al. Regulatory module WRKY33‐ ATL31‐IRT 1 mediates cadmium tolerance in Arabidopsis. Plant,Cell and Environment, 2023, 46(5):1653-1670. |
| [13] |
Ashfaq A, Khan Z I, Ahmad K. Assessing the health risk of cadmium to the local population through consumption of contaminated vegetables grown in municipal solid waste- amended soil. Environmental Monitoring and Assessment, 2022, 194(7):468.
doi: 10.1007/s10661-022-10104-w pmid: 35648302 |
| [14] | Chen J, Yang L B, Yan X X, et al. Zinc-finger transcription factor ZAT 6 positively regulates cadmium tolerance through the glutathione-dependent pathway in Arabidopsis. Plant Physiology, 2016, 171(1):707-719. |
| [15] | Dang F F, Li Y Y, Wang Y F, et al. ZAT10 plays dual roles in cadmium uptake and detoxification in Arabidopsis. Frontiers in Plant Science, 2022, 13:994100. |
| [16] | Dohm J C, Minoche A E, Holtgräwe D, et al. The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature, 2014, 505(7484):546-549. |
| [17] | 王锦霞, 马龙彪, 刘大丽. 甜菜在重金属污染生物修复中的应用潜力. 哈尔滨:中国农业科学院甜菜研究所, 2018:77-80. |
| [18] |
Chen C J, Chen H, Zhang Y, et al. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 2020, 13(8):1194-1202.
doi: S1674-2052(20)30187-8 pmid: 32585190 |
| [19] |
Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 2016, 33(7):1870-1874.
doi: 10.1093/molbev/msw054 pmid: 27004904 |
| [20] | Puentes-Romero A C, González S A, González-Villanueva E, et al. AtZAT4,a C2H2-type zinc-finger transcription factor from Arabidopsis thaliana, is involved in pollen and seed development. Plants-Basel, 2022, 11(15):1974. |
| [21] | Liu Y, Khan A R, Gan Y. C2H 2 zinc-finger proteins response to abiotic stress in plants. International Journal of Molecular Sciences, 2022, 23(5):2730. |
| [22] | Xu G, Guo C, Shan H, et al. Divergence of duplicate genes in exon-intron structure. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(4):1187-1192. |
| [23] |
Yi G, Neelakandan A K, Gontarek B C, et al. The naked endosperm genes encode duplicate indeterminate domain transcription factors required for maize endosperm cell patterning and differentiation. Plant Physiology, 2015, 167(2):443-456.
doi: 10.1104/pp.114.251413 pmid: 25552497 |
| [24] | Seo P J, Ryu J, Kang S K, et al. Modulation of sugar metabolism by an INDETERMINATE DOMAIN transcription factor contributes to photoperiodic flowering in Arabidopsis. The Plant Journal, 2011, 65(3):418-429. |
| [25] | Ingkasuwan P, Netrphan S, Prasitwattanaseree S, et al. Inferring transcriptional gene regulation network of starch metabolism in Arabidopsis thaliana leaves using graphical Gaussian model. BMC Systems Biology, 2012, 6(1):100. |
| [26] |
Coelho C P, Huang P, Lee D, et al. Making roots, shoots, and seeds: IDD gene family diversification in plants. Trends in Plant Science, 2018, 23(1):66-78.
doi: S1360-1385(17)30204-2 pmid: 29056440 |
| [27] |
Kundu A, Das S, Basu S, et al. GhSTOP1,a C2H 2 type zinc- finger transcription factor is essential for aluminum and proton stress tolerance and lateral root initiation in cotton. Plant Biology, 2019, 21(1):35-44.
doi: 10.1111/plb.12895 pmid: 30098101 |
| [28] |
Kobayashi Y, Ohyama Y, Kobayashi Y, et al. STOP2 activates transcription of several genes for Al-and low pH-tolerance that are regulated by STOP1 in Arabidopsis. Molecular Plant, 2014, 7(2):311-322.
doi: 10.1093/mp/sst116 pmid: 23935008 |
| [29] |
El-Sappah A H, Elrys A S, Desoky E M, et al. Comprehensive genome wide identification and expression analysis of MTP gene family in tomato (Solanum lycopersicum) under multiple heavy metal stress. Saudi Journal of Biological Sciences, 2021, 28(12):6946-6956.
doi: 10.1016/j.sjbs.2021.07.073 pmid: 34866994 |
| [30] | 李丹丹. 马铃薯金属耐受蛋白(StMTPs)核心特征分析及StMTP8耐镉功能验证. 贵阳: 贵州大学, 2022. |
| [31] | He X L, Feng T Y, Zhang D Y, et al. Identification and comprehensive analysis of the characteristics and roles of leucine-rich repeat receptor-like protein kinase (LRR-RLK) genes in Sedum alfredii hance responding to cadmium stress. Ecotoxicology and Environmental Safety, 2019, 167:95-106. |
| [32] |
Li H, Li C, Sun D, et al. OsPDR20 is an ABCG metal transporter regulating cadmium accumulation in rice. Journal of Environmental Sciences, 2024, 136:21-34.
doi: 10.1016/j.jes.2022.09.021 pmid: 37923431 |
| [1] | Li Zhi, Guo Xiaoxia, Huang Chunyan, Jian Caiyuan, Tian Lu, Han Kang, Ren Xiaoyun, Ren Huimin, Zhang Peng, Liu Jia, Kong Dejuan, Wang Zhenzhen, Su Wenbin. Effects of Nitrogen Base Fertilizer and Topdressing Ratio on the Growth, Yield and Sugar Content of Sugar Beet under Shallow Buried Drip Irrigation [J]. Crops, 2024, 40(3): 186-191. |
| [2] | Du Hanmei, Tan Lu, Chen Bo, Yu Qiuzhu, Wu Dandan, Wang Anhu. Comprehensive Evaluation of Cadmium Tolerance of Tartary Buckwheat at Seedling Stage [J]. Crops, 2024, 40(2): 40-53. |
| [3] | Lü Baolian, Yang Yuxin, Cui Licao, Shi Feng, Ma Liang, Kong Xiuying, Zhang Lichao, Ni Zhiyong. Identification of bHLH Family Transcription Factors of Wheat and Expression Analysis under Salt Stress [J]. Crops, 2024, 40(1): 65-72. |
| [4] | Zhang Hanwen, Liu Dan, Wang Xuerui, Li Wangshu, Lu Qiang, Wang Shufeng, Zhao Jianan, Wang Yubo, Zhang He, Li Caifeng. Effects of Superimposed Application of BR in Different Periods on Yield and Quality of Sugar Beet under Saline-Alkali Stress [J]. Crops, 2023, 39(4): 237-244. |
| [5] | Yin Xilong, Shi Yang, Li Wangsheng, Xing Wang. Photosynthetic Physiological Response to Drought Stress in Sugar Beet at Seedling Stage [J]. Crops, 2022, 38(6): 152-158. |
| [6] | Li Wangsheng, Wang Xueqian, Yin Xilong, Shi Yang, Liu Dali, Tan Wenbo, Xing Wang. Comprehensive Evaluation of Drought Tolerance of Sugar Beet Germplasms at Seedling Stage [J]. Crops, 2022, 38(6): 54-60. |
| [7] | Du Fu, Xia Maolin, Liu Xinyuan, Yu Zhaojin, Zhang Zhan, Liu Yunfei, Ji Xiaoming. Effective Effects of Acrylamide/Carboxymethyl Cellulose/Biochar Composite Hydrogel on Cadmium Stress in Tobacco Seedlings [J]. Crops, 2022, 38(4): 138-145. |
| [8] | Jiang Yusong, Li Honglei, Li Zhexin, Xu Xiaoyu, Li Longyun, Huang Mengjun. Identification and Expression Analysis of the ZoWRKY Family in Stress Responses Based on Transcriptome Data of Ginger (Zingiber officinale Roscoe) [J]. Crops, 2022, 38(4): 37-45. |
| [9] | Cai Qiqi, Wang Gang, Dong Yinzhuang, Yu Lihua, Wang Yuguang, Geng Gui. Effects of Different Neutral Salt Stress on Photosynthesis and Antioxidant Enzyme System of Sugar Beet Seedlings [J]. Crops, 2022, 38(1): 130-136. |
| [10] | Zhang Ting, Zhang Bowen, Li Guolong, Cao Yang, Li Yue, Zhang Shaoying. Effects of Phosphorus Application Rate and Method on Photosynthetic Performance and Yield of Sugar Beet [J]. Crops, 2021, 37(5): 187-193. |
| [11] | Ding Liuhuizi, Pi Zhi, Wu Zedong. Construction of SSR Fingerprint and Analysis of Genetic Diversity of Sugar Beet Varieties [J]. Crops, 2021, 37(5): 72-78. |
| [12] | Wang Tong, Zhao Xiaodong, Zhen Pingping, Chen Jing, Chen Mingna, Chen Na, Pan Lijuan, Wang Mian, Xu Jing, Yu Shanlin, Chi Xiaoyuan, Zhang Jiancheng. Genome-Wide Identification and Characteristic Analyzation of the TCP Transcription Factors Family in Peanut [J]. Crops, 2021, 37(2): 35-44. |
| [13] | Li Guolong, Wu Haixia, Sun Yaqing. Construction of RNAi Expression Vector of BvWRKY23 Gene in Sugar Beet [J]. Crops, 2020, 36(5): 41-47. |
| [14] | Xu Yuanyuan, Zhao Peng, Hong Quanchun, Zhu Xiaoqin, Pei Dongli. Isolation and Expression Analysis of Transcription Factor Gene TaMYB70 in Wheat [J]. Crops, 2020, 36(4): 84-90. |
| [15] | Zhang Ziqiang, Bai Chen, Zhang Huizhong, Li Xiaodong, Wang Liang, Fu Zengjuan, Zhao Shangmin, E Yuanyuan, Zhang Hui, Zhang Bizhou. Research Progress on Morphology, Physiological and Biochemical Characteristics, and Molecular Level of Salt Stress in Sugar Beet [J]. Crops, 2020, 36(3): 27-33. |
|
||