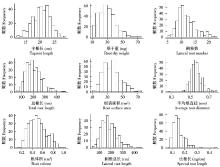
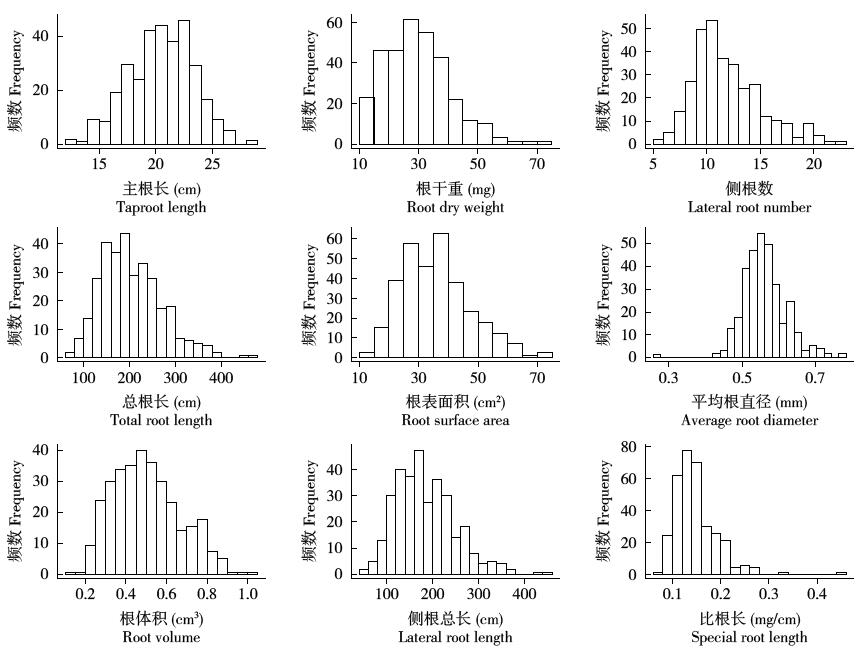
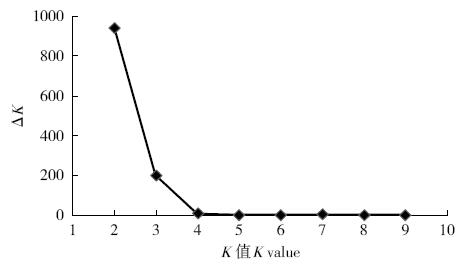
Crops ›› 2019, Vol. 35 ›› Issue (2): 61-70.doi: 10.16035/j.issn.1001-7283.2019.02.009
Previous Articles Next Articles
Association Analysis of Root-Related Traits in Common Bean
Lei Wu,Lanfen Wang,Jing Wu,Shumin Wang
- Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100081, China
| [1] |
Rao I M, Miles J W, Beebe S E , et al. Root adaptations to soils with low fertility and aluminium toxicity. Annals of Botany, 2016,118(4):593-605.
doi: 10.1093/aob/mcw073 pmid: 5055624 |
| [2] |
Rogers E D, Benfey P N . Regulation of plant root system architecture:implications for crop advancement. Current Opinion in Biotechnology, 2015,32:93-98.
doi: 10.1016/j.copbio.2014.11.015 pmid: 25448235 |
| [3] |
Christopher J, Christopher M, Jennings R , et al. QTL for root angle and number in a population developed from bread wheats (Triticum aestivum) with contrasting adaptation to water-limited environments. Theoretical and Applied Genetics, 2013,126(6):1563-1574.
doi: 10.1007/s00122-013-2074-0 pmid: 23525632 |
| [4] |
Hu B, Zhu C, Li F , et al. LEAF TIP NECROSIS1 plays a pivotal role in the regulation of multiple phosphate starvation responses in rice. Plant Physiology, 2011,156(3):1101-1115.
doi: 10.1104/pp.110.170209 pmid: 21317339 |
| [5] |
Ku L X, Sun Z H, Wang C L , et al. QTL mapping and epistasis analysis of brace root traits in maize. Molecular Breeding, 2011,30(2):697-708.
doi: 10.1007/s11032-011-9655-x |
| [6] |
Li R, Han Y, Lv P , et al. Molecular mapping of the brace root traits in sorghum (Sorghum bicolor L. Moench). Breeding Science, 2014,64(2):193-198.
doi: 10.1270/jsbbs.64.193 pmid: 4065327 |
| [7] |
Mace E S, Singh V, Van Oosterom E J ,et al. QTL for nodal root angle in sorghum (Sorghum bicolor L. Moench) co-locate with QTL for traits associated with drought adaptation. Theoretical and Applied Genetics, 2012,124(1):97-109.
doi: 10.1007/s00122-011-1690-9 pmid: 21938475 |
| [8] |
Zheng X, Chen B, Lu G , et al. Overexpression of a NAC transcription factor enhances rice drought and salt tolerance. Biochemical and Biophysical Research Communications, 2009,379(4):985-989.
doi: 10.1016/j.bbrc.2008.12.163 pmid: 19135985 |
| [9] |
Asfaw A, Blair M W . Quantitative trait loci for rooting pattern traits of common beans grown under drought stress versus non-stress conditions. Molecular Breeding, 2011,30(2):681-695.
doi: 10.1007/s11032-011-9654-y |
| [10] |
Beebe S E, Rojas-Pierce M, Yan X , et al. Quantitative trait loci for root architecture traits correlated with phosphorus acquisition in common bean. Crop Science, 2006,46(1):413-423.
doi: 10.2135/cropsci2005.0226 |
| [11] |
Lopez-Marin H D, Rao I M, Blair M W . Quantitative trait loci for root morphology traits under aluminum stress in common bean (Phaseolus vulgaris L.). Theoretical and Applied Genetics, 2009,119(3):449-458.
doi: 10.1007/s00122-009-1051-0 pmid: 19436988 |
| [12] |
Abenavoli M R, Leone M, Sunseri F , et al. Root phenotyping for drought tolerance in bean landraces from Calabria (Italy). Journal of Agronomy and Crop Science, 2016,202(1):1-12.
doi: 10.1111/jac.12124 |
| [13] |
Polania J, Poschenrieder C, Rao I , et al. Root traits and their potential links to plant ideotypes to improve drought resistance in common bean. Theoretical and Experimental Plant Physiology, 2017,29(3):143-154.
doi: 10.1007/s40626-017-0090-1 |
| [14] |
Roman-Aviles B, Snapp S S, Kelly J D . Assessing root traits associated with root rot resistance in common bean. Field Crop Research, 2004,86(2/3):147-156.
doi: 10.1016/j.fcr.2003.08.001 |
| [15] | Sofi P A, Saba I . Natural variation in common bean (Phaseolus vulgaris L.) for root traitsand biomass partitioning under drought. Indian Journal of Agricultural Research, 2016,50(6):604-608. |
| [16] |
Tsuji W, Inanaga S, Araki H , et al. Development and distribution of root system in two grain sorghum cultivars originated from Sudan under drought stress. Plant Production Science, 2005,8(5):553-562.
doi: 10.1626/pps.8.553 |
| [17] | 孙广玉, 何庸, 张荣华 . 大豆根系生长和活性特点的研究. 大豆科学, 1996,15(4):317-321. |
| [18] |
Liao H, Rubio G, Yan X L , et al. Effect of phosphorus availability on basal root shallowness in common bean. Plant Soil, 2001,232(1/2):69-79.
doi: 10.1023/A:1010381919003 pmid: 11729851 |
| [19] |
Armengaud P, Zambaux K, Hills A , et al. EZ-Rhizo:integrated software for the fast and accurate measurement of root system architecture. Plant Journal, 2009,57(5):945-956.
doi: 10.1111/tpj.2009.57.issue-5 |
| [20] |
Li X M, Yuan D J, Wang H T , et al. Increasing cotton genome coverage with polymorphic SSRs as revealed by SSCP. Genome, 2012,55(6):459-470.
doi: 10.1139/g2012-032 pmid: 22670804 |
| [21] |
Chen M L, Wu J, Wang L F , et al. Development of mapped simple sequence repeat markers from common bean (Phaseolus vulgaris L.) based on genome sequences of a Chinese landrace and diversity evaluation. Molecular Breeding, 2014,33(2):489-496.
doi: 10.1007/s11032-013-9949-2 |
| [22] |
Kami J, Velásquez V B, Debouck D G , et al. Identification of presumed ancestral dna sequences of phaseolin in phaseolus vulgaris. Proceedings of the National Academy of Sciences of the United States of America, 1995,92(4):1101-1104.
doi: 10.1073/pnas.92.4.1101 |
| [23] |
Liu K J, Muse S V . PowerMarker:an integrated analysis environment for genetic marker analysis. Bioinformatics, 2005,21(9):2128-2129.
doi: 10.1093/bioinformatics/bti282 pmid: 15705655 |
| [24] |
Evanno G, Regnaut S, Goudet J . Detecting the number of clusters of individuals using the software STRUCTURE:a simulation study. Molecular Ecology, 2005,14(8):2611-2620.
doi: 10.1111/j.1365-294X.2005.02553.x pmid: 15969739 |
| [25] |
Hardy O J, Vekemans X . SPAGEDi:a versatile computer program to analyse spatial genetic structure at the individual or population levels. Molecular Ecology Notes, 2002,2(4):618-620.
doi: 10.1046/j.1471-8286.2002.00305.x |
| [26] |
Bradbury P J, Zhang Z, Kroon D E , et al. TASSEL:software for association mapping of complex traits in diverse samples. Bioinformatics, 2007,23(19):2633-2635.
doi: 10.1093/bioinformatics/btm308 |
| [27] |
Ochoa I E, Blair M W, Lynch J P . QTL Analysis of adventitious root formation in common bean under contrasting phosphorus availability. Crop Science, 2006,46(4):1609-1621.
doi: 10.2135/cropsci2005.12-0446 |
| [28] |
周蓉, 陈海峰, 王贤智 , 等. 大豆幼苗根系性状的QTL分析. 作物学报, 2011,37(7):1151-1158.
doi: 10.3724/SP.J.1006.2011.01151 |
| [29] |
Fang S Q, Yan X L, Liao H . 3D reconstruction and dynamic modeling of root architecture in situ and its application to crop phosphorus research. Plant Journal, 2009,60(6):1096-1108.
doi: 10.1111/tpj.2009.60.issue-6 |
| [30] |
Hund A, Trachsel S, Stamp P . Growth of axile and lateral roots of maize:I development of a phenotying platform. Plant Soil, 2009,325(1/2):335-349.
doi: 10.1007/s11104-009-9984-2 |
| [31] |
Iyer-Pascuzzi A S, Symonova O, Mileyko Y , et al. Imaging and analysis platform for automatic phenotyping and trait ranking of plant root systems. Plant Physiology, 2010,152(3):1148-1157.
doi: 10.1104/pp.109.150748 |
| [32] |
廖红, 严小龙 . 菜豆根构型对低磷胁迫的适应性变化及基因型差异. 植物学报, 2000,42(2):158-163.
doi: 10.3321/j.issn:1672-9072.2000.02.009 |
| [33] |
Pritchard J K, Stephens M, Rosenberg N A , et al. Association mapping in structured populations. American Journal Human Genetics, 2000,67(1):170-181.
doi: 10.1086/302959 pmid: 10827107 |
| [34] |
Zhao K Y, Aranzana M J, Kim S , et al. An Arabidopsis example of association mapping in structured samples. PLoS Genetics, 2007,3(1):e4.
doi: 10.1371/journal.pgen.0030004 pmid: 1779303 |
| [35] |
Gepts P . Biochemical evidence bearing on the domestication of Phaseolus (Fabaceae) beans. Economic Botany, 1990,44(S3):28-38.
doi: 10.1007/BF02860473 |
| [36] |
Zamani Z, Bahar M, Jacques M A , et al. Genetic diversity of the common bacterial blight pathogen of bean,Xanthomonas axonopodis pv. phaseoli,in Iran revealed by rep-PCR and PCR-RFLP analyses. World Journal Microbiology Biotechnology, 2011,27(10):2371-2378.
doi: 10.1007/s11274-011-0705-7 |
| [37] |
Yan X L, Liao H, Beebe S E , et al. QTL mapping of root hair and acid exudation traits and their relationship to phosphorus uptake in common bean. Plant Soil, 2004,265(1/2):17-29.
doi: 10.1007/s11104-005-0693-1 |
| [1] | Xinqi Geng,Huijuan Yang,Yanqing Qin,Xingyou Yang,Shimin Zhao,Hongzhi Shi. Development and Application of Tobacco SSR Markers Based on Genome Re-Sequencing of Different Tobacco Types [J]. Crops, 2019, 35(2): 84-89. |
| [2] | Chunyu Lin,Xiaoyu Liang,Huiyan Zhao,Yang Wang. Analysis of Genetic Diversity and Population Structure of Main Soybean Varieties in Heilongjiang Province [J]. Crops, 2019, 35(2): 78-83. |
| [3] | Zheng Zhang,Yinquan Niu,Dong Zhang,Chengmei Hu,Yichuan Yuan,Huiyan Wang,Shuguang Wang,Yaping Cao,Daizhen Sun. Genome-Wide Association Analysis of Wheat at Heading and Flowering Stages [J]. Crops, 2019, 35(1): 44-49. |
| [4] | Junping He,Shufen Zhang,Jianping Wang,Dongfang Cai,Jinhua Cao,Yancheng Wen,Kun Hu,Lei Zhao,Dongguo Wang,Jiacheng Zhu. Hybrid Purity Identification of Fengyou No.10 by SSR Markers in Bassica napus [J]. Crops, 2019, 35(1): 75-80. |
| [5] | Duan Junjun,Liu Qingfeng,Ren Chunyuan,Yu Song,Yu Lihe,Guo Wei, ,Liang Haiyun. Effects of Different Cultivation Patterns and Densities on Yield and Dry Matter Accumulation of Common Bean [J]. Crops, 2018, 34(6): 110-115. |
| [6] | Tang Liyuan,Li Xinghe,Zhang Sujun,Wang Haitao,Liu Cunjing,Zhang Xiangyun,Zhang Jianhong. QTL Mapping for Photosynthesis Related Traits in Upland Cotton [J]. Crops, 2018, 34(5): 85-90. |
| [7] | Shuai Zhang,Yuhui Pang,Zhenghong Wang,Liming Wang,Chunyan Chen,Zhankui Zeng,Chunping Wang. Variation of Agronomic Traits and Genetic Diversity in Wheat Germplasms [J]. Crops, 2018, 34(2): 44-51. |
| [8] | Lihua Liu,Shaohua Yuan,Shuying Feng,Binshuang Pang,Hongbo Li,Yangna Liu,Liping Zhang,Changping Zhao. Genetic Difference Analysis and Construction of SSR Fingerprinting Database for F-Type Wheat Male Sterile Line and Restorer Lines [J]. Crops, 2017, 33(6): 30-36. |
| [9] | Yunqing Gao,Dongxu Xu,Hongxiao Ren,Qibing Shang,Fang Wang,Cuimian Jiang,Wensheng Huang. Effects of Water and Fertilization Treatments on Water Consumption and Yield of Common Bean [J]. Crops, 2016, 32(6): 124-127. |
| [10] | Sujun Zhang,Liyuan Tang,Cunjing Liu,Zhenxing Jiang,Jina Chi,Haiyan Tian,Xinghe Li,Jianhong Zhang,Xiangyun Zhang. Association Analysis of Fiber Quality with SSR Markers in Gossypium barbadense L. [J]. Crops, 2016, 32(4): 93-100. |
|