Crops ›› 2018, Vol. 34 ›› Issue (5): 85-90.doi: 10.16035/j.issn.1001-7283.2018.05.013
Previous Articles Next Articles
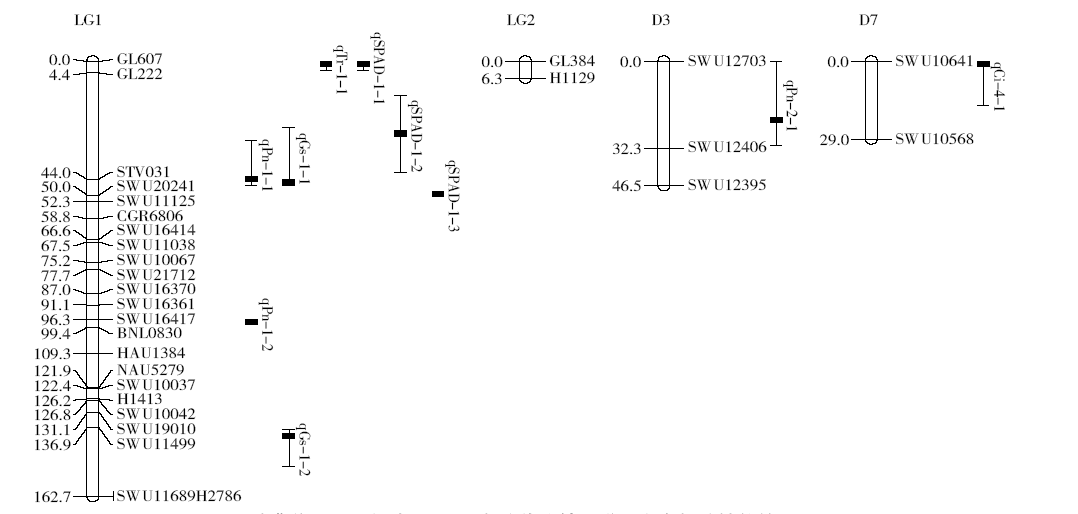
QTL Mapping for Photosynthesis Related Traits in Upland Cotton
Tang Liyuan,Li Xinghe,Zhang Sujun,Wang Haitao,Liu Cunjing,Zhang Xiangyun,Zhang Jianhong
- Institute of Cotton, Hebei Academy of Agriculture and Forestry Sciences/Key Laboratory of Biology and Genetic Improvement of Cotton in Huanghuaihai Semiarid Area, Ministry of Agriculture/National Cotton Improvement Center Hebei Branch, P.R. China, Shijiazhuang 050051, Hebei, China
| [1] | 许晓明, 戴新宾, 陆巍 , 等. 细胞核、细胞质基因与光合作用的关系. 生物学通报, 1999,34(10):5-6. |
| [2] | 耿小红, 武艳芍, 杨林 . 小麦ITMI重组自交系群体的叶片叶绿素含量. 四川农业大学学报, 2017,35(2):139-143. |
| [3] | 刘胜男, 甘剑锋, 张海萍 , 等. 小麦RILs群体叶绿素含量和千粒重相关分析及QTL定位. 安徽农业大学学报, 2013,40(4):570-574. |
| [4] | 余婷婷, 刘朝显, 梅秀鹏 , 等. 玉米光合性状的相关性及QTL分析. 西南大学学报(自然科学版), 2015,37(9):1-10. |
| [5] | 胡茂龙, 王春明, 杨权海 , 等. 水稻光合功能相关性状QTL分析. 遗传学报, 2005,32(8):818-824. |
| [6] | 刘进, 王嘉宇, 姜树昆 , 等. 水稻叶绿素含量动态QTL分析. 植物生理学报, 2012,48(6):577-583. |
| [7] | 李伟, 潘校成, 于洪潇 , 等. 大豆叶绿素含量QTL定位及候选基因预测. 基因组学与应用生物学, 2016,35(7):1793-1799. |
| [8] |
印志同, 宋海娜, 孟凡凡 , 等. 大豆光合气体交换参数的QTL分析. 作物学报, 2010,36(1):92-100.
doi: 10.3724/SP.J.1006.2010.00092 |
| [9] | 戎福喜, 汤丽魁, 唐媛媛 , 等. 海陆渐渗系棉花吐絮期叶绿素含量、荧光参数及相关性状的QTL定位分析. 棉花学报, 2015,27(5):417-426. |
| [10] | 郑巨龙, 龚照龙, 王俊铎 , 等. 新疆陆地棉遗传连锁图谱构建及叶绿素含量和光合速率的QTL定位. 新疆农业科学, 2014,51(9):1577-1582. |
| [11] | 宋美珍 . 短季棉早熟不早衰生化遗传机制及QTL定位. 北京:中国农业科学院, 2006. |
| [12] |
Saranga Y, Menz M, Jiang C X , et al. Genomic dissection of genotype×environment interactions conferring adaptation of cotton to arid conditions. Genome Research, 2001,11:1988-1995.
doi: 10.1101/gr.157201 |
| [13] |
秦鸿德, 张天真 . 棉花叶绿素含量和光合速率的QTL定位. 棉花学报, 2008,20(5):394-398.
doi: 1002-7807(2008)05-0394-05 |
| [14] |
王鹏, 张天真 . 利用棉花海陆种间染色体片段导入系剖析光合色素含量的遗传基础. 作物学报, 2012,38(6):947-953.
doi: 10.3724/SP.J.1006.2012.00947 |
| [15] |
Song X L, Guo W Z, Han Z G , et al. Quantitative trait loci mapping of leaf morphological traits and chlorophyll content in cultivated tetraploid cotton. Journal of Integrative Plant Biology, 2005,47(11) : 1382-1390.
doi: 10.1111/jipb.2005.47.issue-11 |
| [16] | Song X L, Zhang T Z . Molecular mapping of quantitative trait loci controlling chlorophyll content at different developmental stages in tetraploid cotton. Plant Breed, 2010,129:533-540. |
| [17] | 张建 . 陆地棉遗传图谱标记加密与衣分、光合性状的QTL定位. 重庆:西南大学, 2012. |
| [18] | 张建, 刘大军, 林刚 , 等. 陆地棉叶绿素质量分数QTL定位. 西南大学学报(自然科学版), 2011,3(4):1-4. |
| [19] |
Parerson A H, Brubaker C L, Wendel J F . A rapid method for extraction of cotton (Gossypium spp) genomic DNA suitable for RFLP and PCR analysis. Plant Molecular Biology Reporter, 1993,11(2):122-127.
doi: 10.1007/BF02670470 |
| [20] | 张军, 武耀廷, 郭旺真 , 等. 棉花微卫星标记的PAGE/银染快速检测. 棉花学报, 2000,12(5):267-269. |
| [21] |
Zhang T Z, Qian N, Zhu X F , et al. Variations and transmission of QTL alleles for yield and fiber qualities in upland cotton cultivars developed in China. Plos One, 2013,8(2):e57220.
doi: 10.1371/journal.pone.0057220 |
| [22] |
Jamshed M, Jia F, Gong J W , et al. Identification of stable quantitative trait loci (QTLs) for fiber quality traits across multiple environments in Gossypium hirsutum recombinant inbred line population. BMC Genomics, 2016,17:197.
doi: 10.1186/s12864-016-2560-2 |
| [23] |
Liang Z, Lü Y D, Cai C P , et al. Toward allotraploid cotton genome assembly: integration of a high-density molecular genetic linkage map with DNA sequence information. BMC Genomics, 2012,13:539.
doi: 10.1186/1471-2164-13-539 |
| [24] |
Stuber C W, Lincoln S E, Wolff D W , et al. Identification of genetic factors contributing to heterosis in a hybrid from two elite maize inbred lines using molecular markers. Genome, 2006,49(5):545-555.
doi: 10.1139/g06-002 |
| [25] | 陈祖海, 刘金兰, 聂以春 . 陆地棉族系种质系与陆地棉品种间的杂种优势利用研究. 棉花学报, 1994,6(3):151-154. |
| [26] | Bhatt J G , Rao M R K. Heterosis in growth and photosynthetic rates in hybrids of cotton. Euphytica, 1980,30:129-133. |
| [27] |
Li F G, Fan G Y, Lu C R , et al. Genome sequence of cultivated upland cotton (Gossypium hirsutum TM-1). Nature Biotechnology, 2015,33(5):524-530.
doi: 10.1038/nbt.3208 |
| [28] |
Zhang T Z, Hu Y, Jiang W K , et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nature Biotechnology, 2015,33(5):531-537.
doi: 10.1038/nbt.3207 |
| [1] | Wen Zhang,Tao Lu,Yuxia Ye,Quanyi Liu,Yang Feng. Studies on the Effect of Mixed Defoliant on Cotton in Kuitun District, Xinjiang [J]. Crops, 2018, 34(3): 103-107. |
| [2] | Shuai Zhang,Yuhui Pang,Zhenghong Wang,Liming Wang,Chunyan Chen,Zhankui Zeng,Chunping Wang. Variation of Agronomic Traits and Genetic Diversity in Wheat Germplasms [J]. Crops, 2018, 34(2): 44-51. |
| [3] | Lihua Liu,Shaohua Yuan,Shuying Feng,Binshuang Pang,Hongbo Li,Yangna Liu,Liping Zhang,Changping Zhao. Genetic Difference Analysis and Construction of SSR Fingerprinting Database for F-Type Wheat Male Sterile Line and Restorer Lines [J]. Crops, 2017, 33(6): 30-36. |
| [4] | Haihua Luo,Deyi Shao,Gong Chen,Xiumin Xu,Xin Gao,Changkai Yuan,Jinjian Peng,Feiyu Tang. Comparative Analysis of Trait Correlation between Conventional Varieties (Lines) and Hybrids of Cotton [J]. Crops, 2017, 33(5): 31-37. |
| [5] | Changhui Feng,Youchang Zhang,Shu Bie,Jiaohai Zhang,Xiaogang Wang,Songbo Xia,Cheng Zhang,Hongde Qin. Research Progress on Improvement for Verticillium Wilt Resistance by Molecular Marker-Assisted Selection in Cotton [J]. Crops, 2017, 33(5): 21-25. |
| [6] | Min Xu,Yushu Hu,Jinglin Li,Lulu Jin,Zisheng Wang. Clustering and Correlation Analysis of Earlier-Mauture Cotton Innovation Germplasm based on Biological Characters [J]. Crops, 2017, 33(1): 25-31. |
| [7] | Xiefei Zhu,Shenghua Li,Sen Wang. Combining Ability Analysis of Verticillium Wilt-Resistance in Five Resistant Cultivars [J]. Crops, 2016, 32(6): 33-37. |
| [8] | Xue Jiang,Xiaosong Ma,Lijun Luo,Hongyan Liu. QTL Mapping of Phenotypic Traits under Drought Stress Simulated by PEG-6000 in Rice Seedlings [J]. Crops, 2016, 32(5): 31-37. |
| [9] | Sujun Zhang,Liyuan Tang,Cunjing Liu,Zhenxing Jiang,Jina Chi,Haiyan Tian,Xinghe Li,Jianhong Zhang,Xiangyun Zhang. Association Analysis of Fiber Quality with SSR Markers in Gossypium barbadense L. [J]. Crops, 2016, 32(4): 93-100. |
| [10] | Limin Sang,Caifeng Li,Yubo Wang,Lei Liu,Guanghao Guo,Jian Guo,Ming Chen. Effects of Na2SO4+NaCl Mixed Salt Stress on the Growth of Sugar Beet Seedlings [J]. Crops, 2016, 32(4): 137-141. |
| [11] | Tian Weiliang, Ge Zhenhong, Sun Hui. Preliminary Study on Extraction Process of Lignin from Cotton Seed Hulls [J]. Crops, 2013, 29(2): 130-133. |
|